Definition of a critical genetic interval related to kidney abnormalities in the Potocki–Lupski syndrome†‡
The authors have no conflict of interest to declare.
How to Cite this Article: Goh ES-Y, Perez IC, Canales CP, Ruiz P, Agatep R, Yoon G, Chitayat D, Dror Y, Shago M, Goobie S, Sgro M, Walz K, Mendoza-Londono R. 2012. Definition of a critical genetic interval related to kidney abnormalities in the Potocki–Lupski syndrome. Am J Med Genet Part A. 158A:1579–1588.
Abstract
Potocki–Lupski syndrome is a genomic disorder caused by duplication of 17p11.2. It is characterized by failure to thrive, intellectual disability, hypotonia, and behavioral difficulties. Structural renal anomalies have been observed in <10% of affected individuals. We present detailed clinical and molecular data on six patients with Potocki–Lupski syndrome, two of whom had renal abnormalities, and investigate the prevalence of kidney abnormalities in the mouse model for the syndrome. In contrast to affected humans, the mouse model does not demonstrate a renal phenotype. Comparison of the duplicated segment in patients with Potocki–Lupski syndrome and the renal phenotype and the syntenic duplicated region in the mouse model allowed us to suggest a 0.285 Mb critical region, including the FLCN gene that may be important for development of renal abnormalities in patients with this duplication. © 2012 Wiley Periodicals, Inc.
INTRODUCTION
Potocki–Lupski syndrome (PTLS, OMIM 610883) is a genomic disorder caused by duplication of 17p11.2. It is characterized by failure to thrive, intellectual disability, hypotonia, and behavioral difficulties [Potocki et al., 2000, 2007; Treadwell-Deering et al., 2010; Zhang et al., 2010]. The incidence of PTLS is estimated to be 1:20,000, which is in part derived from the frequency of the reciprocal deletion, Smith–Magenis syndrome (SMS, OMIM 182290) [Greenberg et al., 1991; Doco-Fenzy et al., 2008]. Structural renal anomalies have been seen in <10% of patients with PTLS [Zhang et al., 2010] and in 15–35% of individuals with Smith–Magenis syndrome [Greenberg et al., 1996; Potocki et al., 2003].
The common duplicated region on human chromosome 17p11.2 found in patients with PTLS is syntenic to a 32–34 cM region of murine chromosome 11. A mouse model for PTLS, Dp(11)17/+ or Dp/+, that carries a duplication of a region of ∼3 Mb syntenic to the PTLS region was generated through chromosomal engineering [Walz et al., 2003]. Dp/+ mice display most of the clinical characteristics of PTLS, including low body weight, hyperactivity, and social, learning, and memory abnormalities [Walz et al., 2003, 2006; Molina et al., 2008]. Walz et al. [2003] investigated the prevalence of kidney abnormalities in the PTLS mouse model in a mixed C57BL/6 Tyrc-Brd × 129S5/SvEvBrd genetic background. There was no evidence of renal abnormalities on gross pathology or histological examination.
Our study presents a clinical assessment of six patients with PTLS, two of whom have structural renal abnormalities. For comparison, we also investigated the prevalence of kidney phenotype observed in a PTLS mouse model of pure genetic background (C57BL/6-Tyrc-Brd), which we hypothesized could lead to improved understanding of the kidney phenotype in human with PTLS.
MATERIALS AND METHODS
Human Subjects
Patients with duplication 17p11.2 diagnosed by G-banding and FISH studies between January of 2007 and June 2009 were ascertained by one of the participating co-investigators. Informed consent was obtained from the patient's parents and assent obtained when appropriate. The study was approved by the research ethics board at the Hospital for Sick Children, Sickkids Research Institute (Protocol # 1000012127). Each of the patients were assessed by a clinical geneticist (patients 1 and 2 by RML and EG, patients 3–5 by DC, and patient 6 by GY and SG) and their medical records were reviewed.
Cytogenetic and MLPA Analysis
In all cases, G-banding was performed followed by FISH analysis using DAPI-stained nuclei hybridized with probes from the Smith–Magenis syndrome region (17p11.2) in spectrum orange and the RARa locus (17q21) in spectrum green (Probes from Vysis). Multiplex ligation-dependent probe analysis (MLPA) was performed using the MRC Holland SALSA MLPA P064-Mental Retardation 1 kit, which included six probes in the SMS/PTLS region: TNFR-SF13B, RAI1, DFKZP586M1120 (also known as LRRC48), LLGL1, PRPSAP2, and MFAP4 as per manufacturer's instructions using Genemarker (Softgene, State College, PA, Version 1.75).
Animals
The phenotype was analyzed in the C57BL/6-Tyrc-Brd pure genetic background (more than 12 backcrosses to wild-type C57BL/6-Tyrc-Brd). The mice were genotyped visually by the presence of Agouti coat coloration and by PCR. Male and female mice were analyzed in this study. Detailed analysis of histopathology and gross anatomy were conducted as described [Walz et al., 2003]. All procedures were approved by the University of Miami Institutional Animal Care and followed the National Institutes of Health Guidelines, “Using Animals in Intramural Research.”
Statistical Analysis
Statistical analyses were performed by using Student's t-test. A P-value < 0.05 was considered significant.
In Silico Candidate Gene Analysis
After determination of the most informative markers through MLPA, the region of interest was interrogated on the UCSC genome browser using Human hg19 and Mouse mm9 assemblies (http://genome.ucsc.edu/). The candidate genes were cross-referenced to the Online Mendelian Inheritance in Man database (available at http://www.ncbi.nlm.nih.gov/sites/entrez) to determine the tissues of expression where available. If the gene was not an OMIM gene, tissue expression was estimated via the GNF Expression Atlas 2 Data from U133A and GNF1H chips as well as the GNF Expression Atlas 1 Human Data on Affy U95 Chips through the UCSC genome browser (http://genome.ucsc.edu/).
RESULTS
Phenotypic Characterization of Patients With PTLS
All six patients with PTLS were found to have duplication 17p11.2 by G-banding and confirmed by FISH studies. The indications for chromosomal analyses were isolated developmental delay or delay associated with other malformations. Patient 1 was the only one who differed with the clinical indication being neutropenia in the absence of developmental delay. However, family history included a history of cyclic neutropenia on the paternal side. A summary of the clinical characteristics of the six patients can be found in Table I and detailed clinical information for the six patients can be found in Supplemental Table I (Supporting Information online eTable SI). Two of the six patients (patients 1 and 2) were found to have structural renal anomalies.
| Patient | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Sex | F | F | M | M | F | M |
| Age | 1 year, 10 months | 2 years, 5 months | 1 year | 14 years | 1 y 4 mo | 2 y |
| Chromosomal abnormality | dup(17)(p11.2p11.2) | dup(17)(p11.2p11.2) | dup(17)(p11.2p11.2) | dup(17)(p11.2p11.2) | dup(17)(p11.2p11.2) | dup(17)(p11.2p11.2) |
| Duplication size | 2.5 Mb | 1.25 Mb | NR | NR | NR | 3.4 Mb |
| General growth parameters | ||||||
| Low birth weight for gestational age | − | 2,215 g (<3rd centile) | − | 1,810 g (<3rd centile) | − | 2,400 g (<3rd centile) |
| Short stature | − | − | − | 142 cm (<2nd centile) | − | − |
| Microcephaly | − | − | − | 52 cm (∼2nd centile) | − | − |
| Craniofacial features | ||||||
| Broad forehead | + | − | − | + | − | + |
| Downslanted palpebral fissures | − | − | + | − | − | − |
| Epicanthus | − | + | − | + | − | − |
| Overhanging nasal tip | + | − | − | − | − | − |
| Wide nasal bridge | + | + | − | + | + | − |
| Widely spaced eyes | − | − | + | − | − | − |
| Asymmetric smile | + | − | − | − | − | − |
| Retrognathia | − | − | − | − | + | − |
| Medical and developmental history | ||||||
| Poor feeding as an infant | − | + | + | + | − | − |
| Failure to thrive in infancy | − | − | − | + | − | − |
| Hypotonia as an infant | − | + | + | + | + | + |
| Developmental delay | + | + | + | + | + | + |
| Epilepsy | − | − | − | + (febrile) | + (staring) | − |
| EEG abnormalities | − | − | − | NR | NR | − |
| MRI abnormalities | NR | + | NR | NR | NR | + |
| Sleep | ||||||
| Subjective sleep disturbance | − | + | − | NR | − | + |
| Central and/or obstructive sleep apnea | − | NR | − | NR | NR | − |
| Behavior | ||||||
| Autistic featuresa | − | + | + | − | − | + |
| Language impairmentb | − | + | + | + | + | + |
| Systemic | ||||||
| Hypermetropia | − | − | − | − | NR | − |
| Strabismus | − | + | + | − | − | − |
| Hearing impairment | − | NR | + | + | − | − |
| Cardiovascular abnormality | − | − | NR | NR | NR | − |
| Low total cholesterol and low LDL | NR | NR | NR | NR | NR | NR |
| Scoliosis >10° | − | − | − | Kyphosis | − | − |
| Joint laxity | − | − | − | + | − | − |
| Structural renal anomaly | Left multicystic dysplastic kidney | Bilateral hypoplastic kidneys | − | − | − | − |
| Neutropenia | + | − | − | − | − | − |
| Other findings | Left inguinal hernia | Supernumerary nipple, hyperhydrosis, glu-6-phos dehydrogenase deficiency | Mild facial asymmetry with left palpebral fissure being smaller than the right | |||
- NR, not recorded; +, present; −, absent.
- a Autistic features were assessed by the clinician and included poor eye contact, specific sensory needs, stereotypic behaviors and difficulties with social interaction. Details for each patient are described in Supplemental Table SI.
- b Language Impairment was assessed by the clinician based on a developmental assessment expected for their chronological age. Details for each patient appear in Table SI.
Patient 1 had a left multicystic hypoplastic kidney. She was assessed at 22 months of age and did not have other physical findings of PTLS except for mild facial dysmorphisms and some difficulties with speech. She had a 20 word vocabulary but had not started to put two-word sentences together, and her speech was not comprehensible to individuals outside her family. She did not have stereotypic behaviors.
Patient 2 was evaluated at 19 months of age for failure to thrive, developmental delay, left preauricular pit, and bilateral hypoplastic kidneys with increased cortical echogenicity. In addition to these findings, she also had low birth weight, hypotonia, feeding difficulties, abnormal behaviors such as non-purposeful lip smacking movements, and repetitive finger flexions. At 2 years and 5 months of age, she communicated by pointing and simple words and had difficulty interacting with pre-school children her age. Her mother had a bicornuate uterus but no structural or functional renal abnormalities.
MLPA on patient 1 showed a 2.4 Mb region duplicated between markers TNRF-SF13B and MFAP4, while patient 2 had a smaller duplicated region of 1.25 Mb between markers TNFR-SF13B and LLGL1 (Fig. 1).
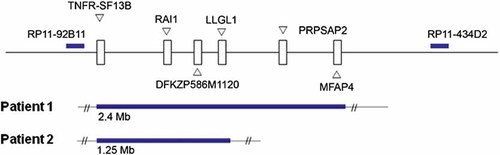
Multiplex ligation-dependent probe analysis of patients 1 and 2 using the six probes in the SMS/PTLS region including TNFR-SF13B, RAI1, DFKZP586M1120 (also known as LRRC48), LLGL1, PRPSAP2, and MFAP4. patient 1 was found to have a 2.4 Mb duplication, while patient 2 had a 1.25 Mb duplication.
Kidney and Urinary Tract Abnormalities Were Evaluated in the PTLS Mouse Model
Kidney function evaluated by urinalysis and blood chemistry showed no significant differences for any genotype, sex, or age groups evaluated (P > 0.05 in all cases), indicating normal functioning renal system. Gross anatomical evaluation of the urogenital system in these mice (N = 38), compared with their wild-type littermates (N = 34) showed normal kidney development in all (Supplemental Data eFigure S1—Supporting Information online). There were no instances of horseshoe kidney, malrotation, ectopia, malposition (right kidney always above left kidney), or unilateral or bilateral bifid ureters (eTable SII—Supporting Information online). Histopathological analysis of the kidneys of Dp/+ and wild-type age matched mice showed no significant differences for the parameters analyzed (Supplemental Data eFigure S1 and eTable SIII—Supporting Information online). A representative image of the histological evaluation for both genotypes can be observed in Supplemental Data eFigure S1 (Supporting Information online).
Comparison of the Duplicated PTLS Genomic Regions in the Human and Mouse
While there are reports of renal abnormalities in humans with PTLS, the mouse model does not show a renal phenotype in either the mixed or pure genetic backgrounds. The syntenic human and mouse regions of PTLS are shown in Figure 2. COPS3 and Zfp179 were the anchoring genes used for making the mouse rearrangements and therefore the PTLS mouse model has duplication of the genes included between them. The gene content in the duplicated region is similar in mice and humans except for a subset of genes which are inside the common duplicated region in patients with PTLS and outside in the mouse due to an inversion in that mouse region of chromosome 11 (Fig. 2—Gray boxes).
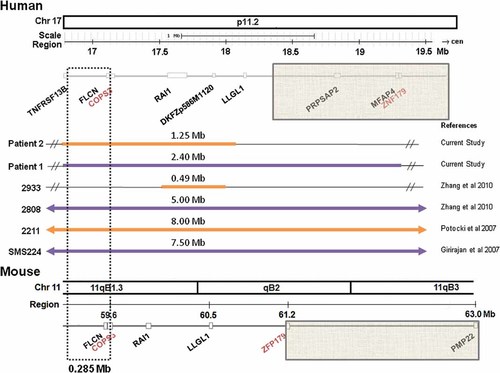
Comparison of human duplications reported with renal phenotypes and the syntenic PTLS mouse region. The genomes were compared using the UCSC genome browser Human February 2009 hg19 and Mouse July 2007 mm9 assemblies (http://genome.ucsc.edu/). Duplicated regions for the individuals in Table III are in the solid, thickened lines. The size of the duplication is represented by the number above the line. COPS3 and ZNF179/ZFP179 were used to make the PTLS rearrangements in the mouse. The areas between the two genes are the regions that are syntenic between human chromosome 17 and mouse chromosome 11. The gray boxes show a large region that is different between the two species. The genes within the dashed lines within a 0.285 Mb region represent candidate genes to explain the discrepancy within the human and mouse phenotype as they are within the region of overlap between the patients with confirmed renal findings and the genes are the same in both mouse and humans with the exception that they are duplicated in the patients and not duplicated in the PTLS mice.
Review of the literature for cases of PTLS with kidney abnormalities identified eight patients with renal abnormalities [Girirajan et al., 2007; Potocki et al., 2007; Zhang et al., 2010]. The present report includes two additional patients bringing the total number to ten. The smallest confirmed duplicated region in a patient with a definite structural renal anomaly is a 1.25 Mb region between TNFR-SF13B and LLGL, as found in patient 2 (Fig. 2). Based on the UCSC genome browser, the 1.25 Mb duplicated region contains 21 genes, 13 of which are listed in OMIM. The names, description and tissue expression of these genes are included in Table II. Within the 1.25 Mb duplicated region in patients with confirmed structural renal anomalies, there is an area that is not duplicated in the PTLS mouse model (Fig. 2—gray-dashed lines). This region is 0.285 Mb and contains six genes (TNFRSF13B, MPRIP, M-RIP, KIAA0864, PLD6, and FLCN) (Table II—gray shading).
| Gene | OMIM number | Description | Function/Clinical significance | Tissue expression |
|---|---|---|---|---|
| TNFRSF13B | 604907 | Tumor necrosis factor receptor superfamily member 13B (Transmembrane activator and CAML interactor) (CD267 antigen). | Common variable immunodeficiency CVID (OMIM#240500) | Spleen, small intestine, thymus, and peripheral blood lymphocytes |
| MPRIP | Myosin phosphatase-Rho interacting protein | Unknown | GNF Microarray expression highest in lymphomas, thyroid and adrenal gland | |
| M-RIP | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | Unknown | GNF Microarray expression highest in parts of the brain and uterus | |
| KIAA0864 | Uncharacterized KIAA0864 protein | Unknown | GNF Microarray expression highest in parts of the brain and uterus | |
| PLD6 | Phospholipase D6 | Unknown | GNF Microarray expression highest expression in lung, liver and ovary | |
| FLCN | 607273 | Folliculin (Birt-Hogg-Dube syndrome protein) (BHD skin lesion fibrofolliculoma protein) | Birt-Hogg-Dube - hair follicle hamartomas, kidney tumors, spontaneous pneumothorax | Most normal adult tissues, including skin, lung, and kidney, and in fetal lung, kidney, liver, and brain tissue |
| COPS3 | 604665 | COP9 signalosome complex subunit 3 (Signalosome subunit 3) (SGN3) (JAB1-containing signalosome subunit 3) | Regions with homology to the 26S proteasome S3 regulatory subunit | cytosolic localization, concentrated around the nucleus |
| NT5M | 605292 | 5'(3')-deoxyribonucleotidase, mitochondrial precursor | Mitochondrial deoxyribonucleotidase DNT2 | Mitochondria |
| MED9 | 609878 | Mediator of RNA polymerase II transcription subunit 9 | promotes activation of RNA polymerase II through direct interactions with transcription factors | GNF Microarray expression highest in thalamus, heart and skeletal muscle |
| RASD1 | 605550 | Dexamethasone-induced Ras-related protein 1 precursor (Activator of G- protein signaling 1) | Produces protein DEXRAS1 expressed in mouse brain, heart, kidney and liver; altered circadian rhythm in mice deleted for Rasd1 | Ubiquitous expression of a 5.0-kb RASD1 transcript, with highest levels in heart |
| PEMT | 602391 | Phosphatidylethanolamine N-methyltransferase | Unknown | PEMT1—in the ER; PEMT2—mitochondria-associated membrane |
| RAI1 | 182290 | Retinoic acid-induced protein 1 | Smith–Magenis syndrome, Chromosome 17p11.2 deletion syndrome | Detected expression in all adult and fetal tissues examined |
| SREBF1 | 184756 | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1) | controls cholesterol homeostasis by stimulating transcription of sterol-regulated genes | The 1a exon predominated in cells that differentiated into adipocytes and in the spleen, whereas the 1c exon predominated in liver cells, white and brown adipose tissue, adrenal gland, and several other tissues of the adult mouse |
| TOM1L | TOM1-like protein 2 (Target of Myb-like protein 2) | Unknown | ubiquitously expressed with higher expression in heart and skeletal muscle | |
| LRRC48 | Leucine-rich repeat-containing protein 48 | Unknown | GNF Microarray expression highest in parts of brain, thyroid, trachea and gonads; some expression in kidney | |
| ATPAF2 | 604273 | ATP synthase mitochondrial F1 complex assembly factor 2, mitochondrial precursor (ATP12 homolog) | Complex V, mitochondrial respiratory chain, deficiency of ATPAF2 subunit | Widely expressed at moderate abundance |
| C17orf39 | Uncharacterized protein C17orf39 | Unknown | GNF Microarray expression highest in testis | |
| DRG2 | 602986 | Developmentally-regulated GTP-binding protein 2 (DRG 2) | Unknown | Northern blot analysis detected DRG2 expression as a major 2-kb and a minor 1.5-kb transcript in various tissues |
| MYO15A | 600316 | Myosin-XV (Unconventional myosin-15) | Deafness, neurosensory, autosomal recessive 3 (non-syndromic) | Northern blot analysis detected MYO15A expression in human fetal and adult brain, and RT-PCR analysis detected expression in human fetal cochlea. RNA dot-blot analysis showed expression in ovary, testis, kidney, and pituitary gland |
| ALKBH5 | Alkylated DNA repair protein alkB homolog 5 | Unknown | GNF Microarray expression highest in testis, heart and kidney | |
| LLGL1 | 600966 | leThal giant larvae homolog 1 (Drosophila) | In Drosophila, lethal giant larvae (Lgl) is essential for asymmetric cortical localization of all basal determinants in mitotic neuroblasts, indispensable for neural fate decisions | Expressed in brain, kidney, and muscle but is barely seen in heart and placenta |
DISCUSSION
Our goal in this study was to compare the renal involvement in humans with PTLS and the PTLS mouse model. In our series, two out of the six patients with PTLS showed a structural renal abnormality, which increases the number of reported renal abnormalities in PTLS to 10. The structural and histopathological renal abnormalities associated with this condition have not been thoroughly investigated. We assessed of the renal phenotype in patients with PTLS and compared them to those found in previously published reports and to the mouse model with this condition to try and delineate the smallest duplicated region associated with renal abnormalities.
Before this report only eight of these patients had kidney abnormalities as presented in Table III [Balarin et al., 1999; Potocki et al., 1999, 2007; Schneider et al., 2000; Girirajan et al., 2007; Zhang et al., 2010]. The size of the duplications in these patients ranged from 0.49 to 8.7 Mb (Fig. 2) [Schneider et al., 2000; Potocki et al., 2007; Zhang et al., 2010]. The patient with the smallest duplication (0.49 Mb) was an 8-year-old male reported with “small kidneys” but they were not structurally or functionally abnormal [L. Potocki, personal communication; Zhang et al., 2010]. Balarin et al. [1999] reported a 19-month-old male with a duplication 17p11.2 (size was not specified) who presented renal electron microscopy abnormalities that were consistent with Alport syndrome. However, he did not have sensorineural hearing loss, ocular abnormalities, nor was there report of whether mutational analyses of COL4A5, COL4A3, or COL4A4 were performed. Surveillance for renal anomalies by renal ultrasound should be considered for all individuals with PTLS as they may be asymptomatic and medical management could change if an abnormality was identified.
| Current study | Current study |
Zhang et al. [2010 ] |
Zhang et al. [2010 ] |
Potocki et al. [2007 ] |
Potocki et al. [2007 ] |
Girirajan et al. [2007 ] |
Schneider et al. [2000 ] |
Potocki et al. [1999 ] |
Balarin et al. [1999 ] |
|
|---|---|---|---|---|---|---|---|---|---|---|
| Patient | Patient 1 | Patient 2 | 2,808 | 2,933 | 1,913 | 2,211 | SMS224 | 1,251 | 1,006 | dup(17) + Alport syndrome |
| Sex | F | F | F | M | F | M | M | F | F | M |
| Age | 1 year, 10 months | 2 years, 5 months | 39 years | 8 years | 13 years | 4 years, 10 months | 8 years | 1 years, 5 months | 14 years | 1 years, 7 months |
| Chromosomal abnormality | dup(17)(p11.2p11.2) | dup(17)(p11.2p11.2) | dup(17)(p11.2p11.2) | dup(17)(p11.2p11.2) | dup(17)(p11.2p11.2) | dup(17)(p11.2p11.2) | dup(17)(p11.2p11.2), 25% mosaicism for tetrasomy 17p11.2p12, 830 kb deletion of 17q11.2q12 | dup(17)(p11.2p11.2) | dup(17)(p11.2p11.2); tandem duplication in prox band p11.2 on one chromosome 17 and deletion in band p12 on the other chromosome 17 | dup(17)(p11.2p11.2) |
| Duplication size | 2.5 Mb | 1.25 Mb | 5 Mb | 0.49 Mb | 3.7 Mb | 8.7 Mb | 7.5 Mb | At least 1.1 Mb | At least 1.1 Mb | NR |
| General growth parameters | ||||||||||
| Low birth weight for gestational age | − | + | − | NR | − | + | NR | + | − | − |
| Short stature | − | − | − | − | + | − | NR | − | +e | − |
| Microcephaly | − | − | − | − | + | − | NR | − | − | − |
| Craniofacial Features | ||||||||||
| Broad forehead | + | − | + | + | + | + | + | + | NR | + |
| Downslanted palpebral fissures | − | − | + | + | + | + | + | + | NR | + |
| Epicanthus | − | + | − | − | NR | NR | − | + | NR | NR |
| Overhanging nasal tip | + | − | + | + | + | + | − | − | NR | NR |
| Broad nasal bridge | + | + | NR | NR | + | NR | − | − | NR | − |
| Widely spaced eyes | − | − | NR | NR | + | − | − | − | NR | − |
| Asymmetric smile | + | − | NR | NR | − | + | − | − | NR | NR |
| Micrognathia/Retrognathia | − | − | + | + | − | − | − | + | + | + |
| Medical and developmental history | ||||||||||
| Poor feeding as an infant | − | + | + | NR | + | + | + | NR | NR | NR |
| Failure to thrive in infancy or early childhood | − | − | + | NR | + | + | − | NR | NR | + |
| Hypotonia as an infant | − | + | NR | NR | + | + | + | − | − | − |
| Developmental delay | + | + | − | + | + | + | + | − (low avg range) | + | +- |
| Epilepsy | − | − | − | − | − | − | + | + (tonic-clonic) | − | − |
| EEG abnormalities | − | − | NR | NR | + | + | + | − | NR | NR |
| MRI abnormalities | NR | + | − | − | + | + | + | − | NR | NR |
| Sleep | ||||||||||
| Subjective sleep disturbance | − | + | − | + | − | + | + | NR | NR | NR |
| Central and/or obstructive sleep apnea | − | NR | NR | NR | + | + | + | NR | NR | NR |
| Behavior | ||||||||||
| Autistic features/hyperactivity | − | + | + | + | + | + | hyperactivity | NR | ADHD | − |
| Language impairment | − | + | − | NR | + | + | + | − | + | − |
| Systems | ||||||||||
| Hypermetropia | − | − | − | − | + | + | Hyperopia + amblyopia | NR | hyperopia | − |
| Strabismus | − | + | − | + | + | NR | − | NR | NR | + |
| Hearing impairment | − | NR | − | − | − | − | + | NR | NR | − |
| Cardiovascular abnormality | − | − | − | − | + | + | + | − | − | NR |
| Low total cholesterol and low LDL | NR | NR | NR | NR | + | − | NR | NR | NR | NR |
| Scoliosis >10 degrees/kyphosis | − | − | − | − | − | − | NR | NR | + | NR |
| Joint laxity | − | − | NR | NR | NR | NR | + | NR | NR | NR |
| Structural renal anomaly | Left multicystic dysplastic kidney | Bilateral hypoplastic kidneys | Mild/mod hydronephrosis suggesting duplication of collecting ducts | − (small kidneys) | − (poor corticomedullary differentiation) | malrotation of L kidney | Malrotation of L kidney | Hypoplastic L kidney | − (episodic microscopic hematuria) | +Electron microscopy consistent with Alport syndrome |
| Neutropenia | + | − | NR | NR | NR | NR | NR | NR | NR | NR |
| Other findings | − | − | − | epistaxis | − | − | Neonatal unconjugated hyperbilirubinemia, thoracolumbar syrinx, recurrent unexplained fevers and leukocytosis | Paternal origin; transient hypoglycemia | Paternal origin; abnormal NCV showing multifocal abnormalities, axonal-type neuropathy | − |
- +, present; −, absence; NR, not recorded; ADHD, attention deficit hyperactivity disorder; NCV, nerve conduction velocities.
Even with a low incidence of kidney abnormalities in patients with PTLS, we expected to find renal or urinary tract anomalies in the Dp/+ mice, but none were identified. Modifier genes outside the duplicated region or variable expressivity in, or different function of, the genes common to the mouse model and the human patients may account for our data. An alternative explanation supported by our data within the 0.285 Mb region highlighted in Figure 2 with gray-dashed lines and Table II in the shaded cells, is that one or more of the genes which are duplicated in the human, and not duplicated in the mouse model, is causative for the genitourinary anomalies in patients with PTLS. We propose that out of the six genes within that region, folliculin (FLCN) is the best candidate as it is the only gene within this region that is expressed in the kidney. Protein truncation mutations in the gene cause the Birt–Hogg–Dube syndrome (OMIM 135150), characterized by renal tumors, cutaneous fibrofolliculomas, and recurrent pneumothoraces or pulmonary cysts. Warren et al. [2004] suggested a role for FLCN as a tumor suppressor by the use of expression studies in normal and neoplastic tissues. Homozygous loss of the Flcn gene in a mouse model displayed early embryonic lethality while their heterozygous counterparts developed kidney cysts and solid tumors with aging [Hasumi et al., 2009]. Moreover, a kidney-specific Flcn knockout mouse model developed enlarged, polycystic kidneys [Chen et al., 2008], further demonstrating the importance of this gene in kidney development. If FLCN is dosage-sensitive and its haploinsufficiency is associated with the formation of renal tumors, it is possible that its overexpression could cause dysplasia or hypoplasia of a normal kidney. Our case series highlights the importance of further studies investigating the role of FLCN in the development of kidney phenotype in patients with PTLS.
Acknowledgements
We thank the patients and their families for their willingness to contribute to learning more about PTLS. This work was in part supported by the Fondation Jerome Lejeune (to KW).




