Genetic and clinical profiles of spondylocostal dysostosis patients in Taiwan† ‡
How to Cite this Article: Wang C-H, Lin W-D, Bau D-T, Chou I-C, Tsai F-J. 2011. Genetic and clinical profiles of spondylocostal dysostosis patients in Taiwan. Am J Med Genet Part A 155: 3132–3135.
Chung-Hsing Wang and Wei-De Lin contributed equally to this work.
To the Editor:
Spondylocostal dysostosis (SCD [OMIM 277300]) comprises a heterogeneous group of disorders characterized by multiple vertebral anomalies, including hemi-vertebrae and segments of vertebrae accompanied by anomalies of the ribs [Bulman et al., 2000]. Patients generally have no family history of the condition and show diverse clinical and radiological findings. However, the basis of these defects in some cases stems from disturbances in the Notch signaling pathway that plays a central role in somite formation [Weinmaster, 2000]. The Notch signaling pathway is an evolutionarily conserved signal-transduction pathway that has important roles throughout embryonic development, as well as in the origins of diseases like cancer [Harper et al., 2003].
In humans, the first autosomal recessive genetic mutation closely related to SCD was identified in the DLL3 gene [Bulman et al., 2000]. Subsequently, three other Notch signaling pathway genes (MESP2, LFNG and HES7) have been associated with SCD [Whittock et al., 2004; Sparrow et al., 2006, 2008]. Mutations in DLL3, include nonsense, missense, frameshift, and splice-site mutations, specify functionally deficient products [Sparrow et al., 1998; Bulman et al., 2000; Bonafe et al., 2003; Turnpenny and Kusumi, 2003]. A genome-wide scanning strategy in one consanguineous family with two affected siblings demonstrated linkage to 15q21.3–15q26.1 and identified a homozygous 4-bp duplication disease-causing mutation in MESP2, which codes for a basic helix-loop-helix transcription factor [Whittock et al., 2004]. A homozygous missense mutation (c.564C → A) in exon 3 of the Lunatic Fringe (LFNG) gene was detected that results in substitution of leucine for phenylalanine, leading to its mislocalization in the cell, dysregulation of Notch signaling, and enzymatic inactivation [Sparrow et al., 2006]. The latest identified SCD sub-type is characterized by shortening of the spine, with multiple and contiguous vertebral segmentation defects involving all spinal regions, but mainly the thoracic spine and ribs anomalies with irregular points of fusion along their length on the right side and very crowded origins on the left. This disorder is called type 4, and involves HES7, which encodes a bHLH-Orange domain transcriptional repressor protein [Davis and Turner, 2001; Sparrow et al., 2008]. The bHLH-Orange domain transcriptional repressor protein not only targets the Notch signaling pathway, it also targets a negative feedback mechanism that attenuates the pathway. These results suggest that proper regulation of the Notch signaling pathway is required for the correct patterning of the axial skeleton [Sparrow et al., 2006, 2008].
In this study, we identified five SCD patients with short stature and congenital scoliosis from four unrelated families were screened for pathogenic mutation(s) in genes of the Notch signaling pathway.
Five Chinese Han patients from four families (three boys and two girls, ranging in age from newborn to 15 years) were diagnosed with SCD by two geneticists with expertise in SCD. Demographic and clinical data for the five patients are presented in Table I. Except for abnormal vertebral segmentation, none of these patients had other systemic anomalies, and all achieved normal developmental milestones. Informed consent to participate in the study was obtained for each subject.
| Age at enrolled (years) | Sex | Body height in cm (percentile) | Body weight in kg (percentile) | Image of spinal CT and/or MRI | Sequencing of DLL3, MESP, LFNG, HES7 genes | |
|---|---|---|---|---|---|---|
| Patient 1a | 15 | F | 131.0 (<3rd) | 39.2 (5–10th) | Decreased size ofvertebral bodies and hypoplasia of disc spaces, and fusion of the posterior neural elements over T3–11 of the thoracic spine | DLL3, 653T → C (Leu218Pro), polymorphism |
| Asymmetric alignment of bilateral ribs | ||||||
| Scoliosis of the T-spine with convexity to | ||||||
| Block vertebra of L4–5 | ||||||
| Patient 2a | 14 | F | 131.1 (<3rd) | 36.3 (5–10th) | Decreased size of vertebral bodies and hypoplasia of disc spaces, and fusion of the posterior neural elements over T3–11 of the thoracic spine | DLL3, 653T → C (Leu218Pro), polymorphism |
| Asymmetric alignment of bilateral ribs | ||||||
| Scoliosis of the T-spine with convexity to right side | ||||||
| Block vertebra of L3–5 | ||||||
| Patient 3 | 8.7 | M | 107.6 (<3rd) | 18.1 (5–10th) | Scoliosis of the T-spine with convexity to right side | DLL3, 653T → C (Leu218Pro) polymorphism |
| Hemivertebra over T2–5 | ||||||
| Absence of 10th ribs | ||||||
| Patient 4 | Newborn | M | 47.2 (<3rd) | 3.1 (25th) | Asymmetric fusion and bifid of ribs | DLL3, IVS4 − 36 C → T, polymorphism LFNG, IVS5 + 68 C → T, polymorphism |
| Butterfly vertebra and hemivertebra over T6, T8–9, and T11 | ||||||
| Scoliosis of T-spine with convexity to right side | ||||||
| Patient 5 | 1.3 | M | 76.2 (<3rd) | 8.0 (<3rd) | Hemivertebra over T10 | DLL3, 515T → G (Phe172Cys), polymorphism |
| Block vertebra of T11–12 | ||||||
| T2–6 partial segmentation anomalies | ||||||
| Scoliosis of the T-spine with convexity to left side |
- F, female; M, male.
- a Patients 1 and 2 are sisters.
Genomic DNA was isolated from peripheral blood using standard DNA extraction techniques. The complete coding regions for DLL3, MESP2, LFNG, and HES7, including the intron/exon boundaries, were screened using previously described methodologies [Bulman et al., 2000; Whittock et al., 2004; Sparrow et al., 2006, 2008].
Amplified PCR products were purified from agarose gel using QIAEX II (Qiagen, Hilden, Germany) and subjected to direct sequencing for mutation identification. Direct sequencing was performed using the BigDye 3.1 Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) with the ABI 3100 Genetic Analyzer (Applied Biosystems). Sequencing data were collected at least twice to optimize accuracy and representation.
Clinically, all five patients were below the 3rd centile for height relative to the reference growth chart obtained from the Department of Health in Taiwan. Obvious scoliosis and short stature were attributed to abnormal development and malalignment of their spines. Among the participants, Patients 1 and 2 are sisters and exhibit marked phenotypic similarity with decreased size of vertebral bodies, hypoplasia of disc spaces through the thoracic spine, T-spine scoliosis, and block lumbar vertebra. The other three unrelated patients displayed varying degrees of block and hemi-vertebra in the thoracic spine, with asymmetric fusion, bifid ribs, and thoracic butterfly vertebra. Radiographs showed failure of normal vertebral segmentation and are presented in Figure 1, with the exception of Patient 4.
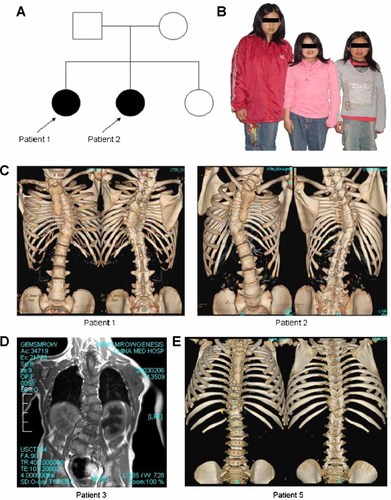
A: The pedigree of the two representative patients (i.e., Patients 1 and 2). B: Patients 1 and 2 show short stature (with shortened trunk) and congenital scoliosis while their younger sister (left) shows normal stature. C: Multiple vertebral anomalies and asymmetric alignment of ribs in the chest cage and block lumbar spines, observed in reconstructed 3D CT images. D,E: Diverse abnormal image characteristics in vertebral segmentation and rib alignment from Patients 3 and 5.
Direct sequencing identified four specific single nucleotide variants in five patients (Table I). Among them, three of the sequence variants were located in DLL3—515T → G (Phe172Cys), 653T → C (Leu218Pro), and IVS4 − 36 C → T. The fourth single nucleotide variant was located in LFNG: IVS5 + 68 C→T. The first two sequence variants were situated within coding regions of the DLL3 gene, and, according to a previous report, were deemed as polymorphic markers that had no association with SCD type 1 phenotype [Sparrow et al., 2002]. The remaining two intronic polymorphisms were located deep in the intron areas of their respective genes, which make them unlikely to be disease-causing.
For this study, we initially recruited 10 patients who had presented with scoliosis and short stature; but only those with non-progressive scoliosis, no other systemic structure anomalies, no neurological deficits, and normal mentality on subsequent follow-up were included in this analysis. Ultimately, this left us with five phenotypically heterogeneous patients with abnormal vertebral segmentation. However, none of our patients completely fulfilled the criteria for the strictest definition of SCD, which requires contiguous involvement of at least 10 spinal segments and aberrant rib alignment, with some asymmetry in rib alignment and irregular points of rib fusions, but basic overall symmetry of thorax shape. It is possible that in addition to the four published sub-types of SCD that have been identified, other sub-types of SCD, may exist in the Taiwanese population. This possibility may explain why we failed to identify mutations in these SCD patients by candidate genes approach of four known SCD responsible genes.
In a knockout mouse study, a targeted mutation of Dll1 (a component of the mouse Notch signaling pathway) was identified and led to a severe decrease or absence of expression of genes in the Notch signaling pathway including Mesp2 [Kusumi et al., 2004]. On the other hand, mutations in Dll3 may have effects not only on Mesp2, but also its downstream genes, Lfng, Hes1, and Hes5 [Kusumi et al., 2004]. In addition, genes not so closely related to the Notch signaling pathway, like GDF6, FGF8, and WNT, also are involved in the etiology of somitogenesis and associated with some specific clinical phenotypes and diseases. These data support the notion that somitogenesis is not regulated solely by the four well-known genes DLL3, MESP2, LFNG, and HES7 [Aulehla et al., 2003; Tassabehji et al., 2008; Dubrulle et al., 2011].
Among these five cases, two sisters (Patients 1 and 2) were notable as they exhibited the greatest degree of similarity genetic genotype background, age, and disease appearance. Their parents are descendants of the same minor aboriginal tribe in Taiwan; and, even though they came from different mountain areas in the central territory of this island country, the possibility of distant consanguinity cannot be excluded. The mode of inheritance is likely autosomal recessive. Their radiographic findings included less severe shortening of the spine than the four established sub-types of SCD. Analysis of haplotypes and other candidate genes is warranted in the Taiwan population with characteristics of SCD.
Acknowledgements
We thank Wen-Shin Chang, Yu-Lin Liao, and Judy Wang for their technical assistance. This study was supported by research grants and China Medical University and Hospital (CMU99-NTU-10).




