Holoprosencephaly in a family segregating novel variants in ZIC2 and GLI2 †
How to Cite this Article: Wannasilp N, Solomon BD, Warren-Mora N, Clegg NJ, Delgado MR, Lacbawan F, Hu P, Winder TL, Roessler E, Muenke M. 2011. Holoprosencephaly in a family segregating novel variants in ZIC2 and GLI2. Am J Med Genet Part A 155:860–864.
Abstract
Holoprosencephaly (HPE) is the most common malformation of the human forebrain. Typical manifestations in affected patients include a characteristic pattern of structural brain and craniofacial anomalies. HPE may be caused by mutations in over 10 identified genes; the inheritance is traditionally viewed as autosomal dominant with highly variable expressivity and incomplete penetrance. We present the description of a family simultaneously segregating two novel variants in the HPE-associated genes, ZIC2 and GLI2, as well as the results of extensive population-based studies of the variant region in GLI2. This is the first time that multiple HPE-associated variants in these genes have been reported in one family, and raises important questions about how clinicians and researchers should view the inheritance of conditions such as HPE. © 2011 Wiley-Liss, Inc.
INTRODUCTION
Holoprosencephaly (HPE), the most common malformation of the human forebrain, may be due to diverse genetic and teratogenic causes, including mutations in over 10 genes [reviewed in Pineda-Alvarez et al., 2010; Roessler and Muenke, 2010]. Mutations in SHH, ZIC2, and SIX3 together account for approximately 20% of non-chromosomal, non-syndromic HPE, while mutations in less commonly associated genes, such as PTC1, TGIF, and GLI2 are estimated to account for up to 2% of cases together [Roessler et al., 1996, 2003, 2005; Brown et al., 1998; Wallis et al., 1999; Gripp et al., 2000; Ming et al., 2002; reviewed in Pineda-Alvarez et al., 2010]. Inheritance is autosomal dominant, with incomplete penetrance and highly variable expressivity, ranging from severe brain anomalies incompatible with life to subtle facial findings without structural brain anomalies (sometimes termed “microform” HPE) [Solomon et al., 2010b].
Though a satisfactory explanation of the wide degree of severity in mutation-positive individuals is far from complete, the current hypothesis hinges on multiple interacting genetic and environmental factors [Ming and Muenke, 2002; reviewed in Monuki, 2007; Schachter and Krauss, 2008; Roessler and Muenke, 2010]. We present an unusual case of a family found to segregate mutations in two HPE-associated genes, ZIC2 and GLI2. ZIC2 encodes a transcription factor involved in both axial midline establishment and in the development of the dorsal telencephalon [Cheng et al., 2006; Warr et al., 2008]. GLI2 encodes a transcription factor that acts as an obligatory mediator of Sonic Hedgehog function in the patterning of the ventral forebrain [Roessler et al., 2003, 2005]. This is the first time in one family that variants in multiple HPE-associated genes have been described in these genes, and this example may increase our understanding of the complex pathogenesis of HPE. Specifically, classical HPE has previously been associated with diminished ventral hedgehog signaling and a second (previously thought to be distinct) type of HPE, the middle hemispheric variant (MIHV) type, has been suggested to result from mis-patterning related to disturbances in dorsal acting factors including Wnts, BMPs, and possibly ZIC2 [reviewed in Monuki, 2007; Fernandes and Hébert, 2008].
FAMILY DESCRIPTION
The proband is a 7-year-old African-American female with semilobar HPE (Fig. 1). Additional clinical findings include severe, global developmental delay, diabetes insipidus, and dysmorphic craniofacial features consisting of microcephaly, bitemporal narrowing, upslanting palpebral fissures, a flat nasal bridge, a short nose with anteverted nares, a broad and deep philtrum, and a high palate. Routine chromosome analysis revealed no abnormalities. As part of a standard panel testing the four genes most commonly associated with HPE, bidirectional sequencing was performed for mutations in SHH, ZIC2, SIX3, and TGIF [methods described in Roessler et al., 1996; Brown et al., 1998; Wallis et al., 1999; Gripp et al., 2000], which revealed a base change predicted to cause a premature truncation in the ZIC2 protein: c.1206C>G, resulting in p.Tyr402X (RefSeq ID for ZIC2: NM_007129.2). This mutation was subsequently found in the patient's mother, who did not have a previously confirmed diagnosis of HPE, but who had mild developmental delay (no neuroimaging has been performed). The patient's brother, who was believed to have cerebral palsy ascribed to a possible neonatal stroke (a neonatal sonogram did not show signs of HPE, but no further neuroimaging is available) was also found to have the same mutation in ZIC2. Neither the mother nor the brother were described as having any dysmorphic facial features typical of HPE or other signs of microform HPE, though further details for the family are not available.
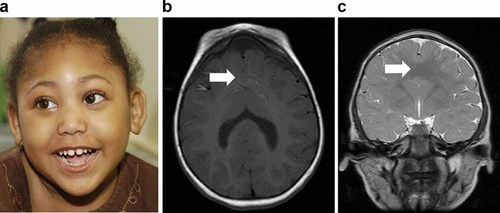
a: Facial view of the proband (shown with consent), a 7-year-old female with semilobar HPE and variants in both GLI2 and ZIC2. The craniofacial phenotype in this patient, which includes microcephaly, bitemporal narrowing, upslanting palpebral fissures, a flat nasal bridge, a short nose with anteverted nares, a broad and deep philtrum, and a high-arched palate, is similar to other patients with mutations in ZIC2 [Solomon et al., 2010a]. b,c: Neuroimaging of the proband demonstrates semilobar HPE, with a clear lack of separation, indicated by white arrows, of the anterior hemispheres, as shown on axial (b) and coronal (c) sections.
Subsequently, sequencing of GLI2 on a research basis [methods described in Roessler et al., 2003, 2005] revealed a novel change in the proband: c.677G>A, resulting in p.Arg226His (RefSeq ID for GLI2: NM_005270). This variant was also found in the proband's mother and brother (Fig. 2). Though this nucleotide change results in the substitution of one basic polar amino acid for another, the amino acid is located at a highly evolutionarily conserved residue in a motif that itself has been recognized to be conserved from flies to humans over 500 million years of evolution (Supplemental Figure). Furthermore, this variant was not found in 928 chromosomes of normal controls by TaqMan assay, including 738 chromosomes from Caucasian individuals and 190 chromosomes from ethnically matched (African-American) individuals (Supplemental Table). Additionally, bidirectional sequencing of the amplicon containing the variant in 1096 chromosomes from normal controls, including 914 chromosomes from Caucasian individuals and 182 chromosomes from African-American individuals, did not reveal any control individuals with this variant. (A nearby variant in the GLI2 found in another, unrelated individual with phenotypic findings consistent with a possible mutation in an HPE-associated gene, c.686G>T, resulting in p.Arg229Leu, was found in 2 of 1096 chromosomes from normal controls, both of which were from Caucasian individuals; a c.691C>T (p.Arg231Trp) variant found in a child with HPE (unknown type) was not found in any control samples.)
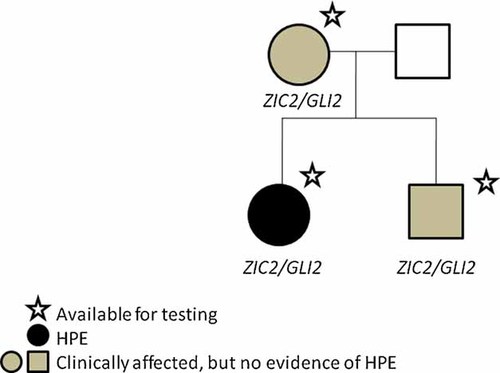
Pedigree of the described family. Individuals with stars were available for testing. Genes with identified variants are indicated beneath each individual.
DISCUSSION
This report focuses on proposed digenic inheritance in HPE, in which multiple genetic factors (in this case in GLI2 and ZIC2) act in combination to yield a disease phenotype. Of the approximately 1,000 patients with HPE described in the medical literature, 6 patients with multiple variants in HPE-associated genes have been previously reported (Table I) [Nanni et al., 1999; Ming and Muenke, 2002; El-Jaick et al., 2007; Lacbawan et al., 2009; Solomon et al., 2010a]. Proving the pathogenicity of these variants can be extremely challenging, and using such results to direct genetic counseling is an intricate process. In the family described here, the mutation in ZIC2, which results in a protein truncation, is certain to be loss-of-function, and is expected to have phenotypic consequences [Roessler et al., 2009b]. Supporting this is the fact that the proband's facial appearance is very typical of patients with mutations in ZIC2 [Solomon et al., 2010a].
| Patient | Clinical information (for all individuals segregating variants) | Genes/variants | Functional data | Refs. |
|---|---|---|---|---|
| 1 | Female with semilobar HPE; phenotypically normal mother | SHH: c.1270C>G, p.Pro424Ala (maternal); TGIF: del18p11 (due to maternal balanced translocation) | SHH: likely functionally normal allele; TGIF: deletion |
Nanni et al. [1999 ], Ming and Muenke [2002 ], Roessler et al. [2009a ] |
| 2 | Female with HPE (type unknown); phenotypically normal mother | SHH: c.1132_1140del, p.378_380del (maternal); TGIF: c.451A>G, p.Thr151Ala (not maternal; paternal testing not available) | SHH: predicted loss-of-function; TGIF: likely functionally normal allele |
Nanni et al. [1999 ], Ming and Muenke [2002 ], El-Jaick et al. [2007 ], Roessler et al. [2009a ] |
| 3 | Female with HPE (type unknown) | SHH: c.869G>A, p.G290Asp; ZIC2: c.1377_1406dup30, p.A461_470dup; inheritance of both mutations unknown | SHH: likely functionally normal allele; ZIC2: severe loss-of-function |
Nanni et al. [1999 ], Ming and Muenke [2002 ], Roessler et al. [2009a ,b ], Solomon et al. [2010a ] |
| 4 | Male with semilobar HPE; phenotypically normal father | PTC1: c.1165G>A, p.Ala393Thr (paternal); SIX3: c.278C>A, in p.Ala93Asp (de novo) | PTC1: in functionally important, conserved region; SIX3: severe loss-of-function |
Ming et al., [2002 ], Domené et al. [2008 ], Lacbawan et al. [2009 ] |
| 5 | Female with semilobar HPE; both parents phenotypically normal except for hypothyroidism | TGIF: c.228C>A, p.His76Gln (maternal); TGIF: c.140_141delTG, p.Val47AlafsX9 (paternal) | TGIF p.His76Gln: likely functionally normal allele; TGIF p.Val47AlafsX9: truncation |
El-Jaick et al. [2007 ] |
| 6 | Female with semilobar HPE | SIX3: c.850G>C, p.Ala284Pro; ZIC2 (c.910T>C), p.Trp304Arg; Inheritance of both mutations unknown | SIX3: unknown; ZIC2: predicted null |
Lacbawan et al. [2009 ], Roessler et al. [2009b ], Solomon et al. [2010a ] |
- As shown, evidence regarding the pathogenicity of each variant is highly variable.
The functional result of the variant in GLI2 is harder to interpret. While this variant is located in a highly evolutionarily conserved region and was not found in over 1,000 control chromosomes (including ethnically matched individuals), no members of the family appear to have findings typical of patients with mutations in GLI2. Though relatively few individuals with bona fide loss-of-function mutations in GLI2 have been reported, patients tend to have more severe perturbations of endocrinological function than isolated posterior pituitary insufficiency and may display postaxial polydactyly [Roessler et al., 2003, 2005; Solomon et al., 2010b]. As none of these relatively GLI2-specific findings was present in any members of this family, the GLI2 variant may indeed be simply a rare familial variant not associated with disease.
Predicting the hypothetical consequences of the identified GLI2 variant is also challenging, as the function of the region in which this particular variant is located is not completely understood. Much of the regulation of GLI2 function involves proteosomal degradation [Huntzicker et al., 2006; Zhang et al., 2009], and the amino acid change in this family is located in the GLI2 NR/Degron N domain, the region that also binds to the GLI2 repressor SUFU [Murone et al., 2000; Cheng and Bishop, 2002; Barnfield et al., 2005; Croker et al., 2006; reviewed in Tsanev et al., 2009]. Possible mechanisms by which the GLI2 variant could result in pathogenicity include destabilization of GLI2 function (which would be predicted to result in an HPE-like phenotype). We can also envision different alterations leading to inhibition of proteosomal degradation, or atypical stabilization of either activator or repressor forms similar to some of the described disorders of GLI3 function [Kang et al., 1997; Roessler et al., 2003, 2005; Johnston et al., 2005]. Ultimately, further answers will rest on the results of functional assays and assessment of additional patients for similar genetic changes. Complex interactions between networks of Gli genes and interacting elements makes interpretation of even the most sophisticated functional assays challenging; ultimately, clinical studies in humans may yield results that are as important as from any other type of scientific inquiry [Chen et al., 2009].
In summary, this family is the first in which variants in both of these HPE-associated genes have been identified. The results, which raise difficult scientific and clinical counseling issues, clearly highlight some of the challenges of studying a complex neurological disorder such as HPE. In terms of practical considerations, families such as that described here emphasize the need to conduct testing of more than one HPE gene in affected families, and that multiple testing modalities are required, including both sequence-based and copy-number based modalities. At a minimum, our group recommends an initial karotype, followed by sequence based familial testing of the most commonly associated HPE-loci (SHH, ZIC2, and SIX3), with further testing according to phenotypic considerations. Gene sequencing should be performed in conjunction with testing designed to look for genomic imbalances (such as array comparative genomic hybridization) [Pineda-Alvarez et al., 2010].
Acknowledgements
This research was supported by the Division of Intramural Research, National Human Genome Research Institute, National Institutes of Health, Department of Health and Human Services, United States of America. The clinical research was supported by the Don and Linda Carter Foundation and the Crowley-Carter Foundation.




