Evidence for a recurrent microdeletion at chromosome 16p11.2 associated with congenital anomalies of the kidney and urinary tract (CAKUT) and Hirschsprung disease†
How to Cite this Article: Sampson MG, Coughlin CR, Kaplan P, Conlin LK, Meyers KEC, Zackai EH, Spinner NB, Copelovitch L. 2010. Evidence for a recurrent microdeletion at chromosome 16p11.2 associated with congenital anomalies of the kidney and urinary tract (CAKUT) and Hirschsprung disease. Am J Med Genet Part A 152:2618–2622.
Abstract
Congenital Anomalies of the Kidney and Urinary Tract can be associated with Hirschsprung disease. We report on three children with a similar 16p11.2 microdeletion with a spectrum of clinical anomalies consisting of congenital anomalies of the kidney and urinary tract in two patients (Patients 1 and 2) and Hirschsprung disease in two patients (Patients 1 and 3), leading us to hypothesize that a gene in this region is associated with these phenotypes. Patient 1 presented with left renal agenesis, grade-IV vesicoureteral reflux, and Hirschsprung disease, Patient 2 with left renal agenesis, chronic kidney disease, chronic constipation, seizures, and developmental delay, and Patient 3 with Hirschsprung disease and normal kidneys. Genome-wide microarray analysis demonstrated overlapping microdeletions within 16p11.2. The shortest region of overlap in the three patients contained only eight genes, including the SH2 domain-containing binding protein 1 (SH2B1), an adaptor protein which has been implicated in enhancement of the tyrosine kinase activity of RET, whose role in developmental disease of the kidney and enteric enervation is well established. Our findings suggest that 16p11.2 deletions are associated with abnormalities of renal and enteric development and we hypothesize that deletion of SH2B1 may account for the observed phenotype. © 2010 Wiley-Liss, Inc.
INTRODUCTION
The association of congenital anomalies of the kidney and urinary tract (CAKUT) and Hirschsprung disease (HSCR) has been appreciated for decades [Sinnassamy et al., 1986; Amiel and Lyonnet, 2001; Moore, 2006]. The discovery that mice with deletions of REarranged after Transfection (RET) [Schuchardt et al., 1994] or Glial Derived Neurotrophic Factor (GDNF) [Moore et al., 1996] genes had abnormalities in renal morphogenesis and enteric enervation served to implicate the RET–GDNF signaling pathway in this process. It is estimated that 50% of familial and 7–35% of patients with HSCR result from RET mutations [Moore and Zaahl, 2008]. More recently, there have been reports of RET mutations in simplex patients with CAKUT without abnormalities of enteric enervation [Skinner et al., 2008; Mace et al., 2009].
There is evidence that site-specific mutations within the RET gene result in variability of both the CAKUT and HSCR phenotypes [Kashuk et al., 2005; Skinner et al., 2008; Jain et al., 2010]. However, in the largest study describing this association, 25% of those with HSCR (21/84 patients) had a CAKUT phenotype but only 1/21 of these patients had a mutation in RET [Pini Prato et al., 2009]. In addition, sequencing of GDNF and GDNF Family Receptor Alpha 1 (GFRα1), other key members of the RET–GDNF pathway, in these patients did not show any mutations [Pini Prato et al., 2009].
From these studies, it is clear that abnormalities in genes other than RET and GDNF result in abnormalities of both renal morphogenesis and enteric enervation. Given the central role of the RET–GDNF signaling in these processes, it is plausible that other genes involved in this pathway could be implicated. We report three patients with microdeletions at 16p11.2 who presented with a spectrum of the CAKUT–HSCR phenotype. We studied these deletions to identify candidate gene(s) within the shortest region of overlap that could explain the observed phenotypes.
CLINICAL REPORTS
Patient 1
A 2-day-old male was evaluated for abdominal distention and bilious emesis. A 20-week prenatal ultrasound showed left renal agenesis. Gestational age was 40 weeks. His mother and grandmother had frequent urinary tract infections. There was no known history of renal or gastroenterological abnormalities. His length was 50 cm (∼75th centile), weight 3.49 kg (25th–50th centile), and head circumference 34 cm (∼10th centile). Blood pressure was 85/59. He was not dysmorphic. His ears were normal without preauricular pits or tags. There were no branchial clefts or cysts. The cardiac exam was normal. The abdomen was distended without visceromegaly. External genitalia were normal. The anus was patent. A rectal suction biopsy demonstrated absent ganglion cells. The aganglionic bowel segment extended to the mid-transverse colon. A colostomy was performed. A renal ultrasound revealed left renal agenesis and a normal sized (5.3 cm) right kidney with normal cortical echogenicity, corticomedullary differentiation, but no hydronephrosis. There was mild increased medullary pyramid echogenicity. A voiding cystourethrogram demonstrated grade 4 reflux, with dilation and tortuosity of the ureter, pelvicaliectasis, and a normal urethra. His creatinine was 0.6 mg/dl at 24 days of age.
Patient 2
A 17-year-old boy had left renal agenesis, chronic kidney disease (CKD) stage 2, proteinuria, hyperuricemia, chronic constipation, seizure disorder, migraine headaches, pervasive developmental disorder, retinal dystrophy, retinitis pigmentosa, and kyphosis. A maternal great uncle had gout and arthritis. There was no family history of any other renal or gastroenterologic disorders. At 17 years, his height was 178.1 cm (50–75th centile), weight was 75.5 kg (∼50th centile), and BMI was 24 kg/m2 (∼75th centile). He had a long oval face, with a low anterior hairline, high nasal bridge, and small mouth with full lips. His cardiac, pulmonary, and abdominal exams were normal. He was hypotonic with global developmental delay. He had severe kyphosis, decreased supination of the forearms, bilateral fifth finger clinodactyly, and small joint hypermobility.
A renal ultrasound was last performed in 2004 and showed a single right kidney of 12 cm in length (>95th centile) with normal echogenicity and corticomedullary differentiation. He excreted 2.2 g of protein per day. He was treated with an angiotensin receptor blocker for treatment of proteinuria. His current serum creatinine is 1.5 mg/dl (estimated glomerular filtration rate of 83 ml/min/m2). He is treated with allopurinol for hyperuricemia, with a current serum level of 6.6 mg/dl. An upper GI study was normal; upper endoscopy showed mild esophagitis, but there were no abnormalities on esophageal, antral, or duodenal biopsy. He did not have a rectal biopsy. He takes polyethylene glycol, lactulose, and intermittent enemas for constipation.
Patient 3
A 12-year-old boy was evaluated at 2 days of age as a result of abdominal distention and obstipation. His prenatal course was uncomplicated. Gestational age was 41 weeks; birth weight was 2.9 kg (∼25th centile), and length 53.3 cm (∼90th centile). He had congenital contractures of the metacarpophalangeal, proximal interphalangeal, and distal interphalangeal joints of the fingers and toes, and elbows, mild crumpling of the ear helices but no ear pits or tags, and right intra-abdominal testis. The patient's father, paternal grandmother and one paternal cousin were reported to have contractures. The patient was diagnosed with Beals syndrome (congenital contractural arachnodactyly, OMIM# 12150) based on the clinical findings and family history. DNA sequencing of FBN2 (the gene which encodes fibrillin-2 and in which mutations result in Beals syndrome) showed a heterozygous c.3777T>A mutation, which predicts p.Asn1259Lys, which is a conserved amino acid in the calcium binding epidermal growth factor like domain 15. This is consistent with a disease-causing mutation in FBN2 and consistent with the clinical diagnosis.
There was no family history of renal abnormalities or gastroenterological problems. A barium enema and recto-sigmoid biopsies were consistent with short-segment HSCR. A Duhamel pull-through procedure and orchiopexy were done at 2 weeks of age. At 12 years, his BMI was 18.3 kg/m2 (∼50th centile). His blood pressure and estimated glomerular filtration rate were normal. Renal ultrasound was normal. He had a history of an auditory processing and attention deficit hyperactivity disorder.
MATERIALS AND METHODS
For each patient and parental sample, DNA was extracted from peripheral blood and quality was monitored by analysis of OD260/OD280 and OD260/OD230 ratios. Acceptable samples had values between 1.8 and 2.0 and ratios > 2.0, respectively. Thirty microliters of a 100 ng/µl solution of genomic DNA was genotyped on the Illumina BeadStation (Illumina, Inc., San Diego, CA). A genome-wide single nucleotide polymorphism (SNP) array analysis was performed using the Illumina Quad610 array and BeadStation Scanner and Bead Studio analysis software (Illumina, Inc.) to detect pathologic deletions and duplications. In preparation for analysis, the samples were whole genome-amplified, fragmented, hybridized, fluorescently tagged and scanned, as per standard protocols [Gunderson et al., 2005]. Analysis was carried out using the Beadstudio Genotyping module and DNA copy number changes were determined using B-allele frequency and log 2 R ratio [Gai et al., 2010]. The nucleotide position of the first and last SNP deleted for each patient was reported based upon the UCSC Genome Browser Build 36/hg18, Mar. 2006 assembly. Parental genotyping information was used to verify paternity and to determine inheritance for the 16p11.2 deletion identified in Patients 1 and 2.
RESULTS
The genome-wide SNP array analysis showed a microdeletion at 16p11.2 for Patients 1, 2, and 3 (Fig. 1). Patients 2 and 3 had identical 1.69 Mb deletions at 16p11.2 (minimal deletion region: nucleotide 28,396,413–30,085,308, maximal deletion region: 28,249,997–30,239,704). Both boys also had pervasive developmental disorder, auditory processing difficulties, and attention deficit hyperactivity disorder. The deletion in Patient 2 was found to be de novo, however parental samples were not available for Patient 3.
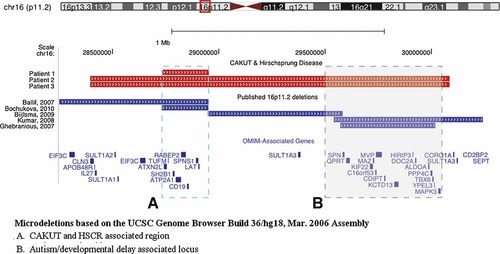
Microdeletions at 16p11.2. Graphical representation of the chromosome 16p11.2 deletions discovered by genome-wide single nucleotide polymorphism array analysis in the three patients reported (red rectangles) and a comparison to previously reported deletions in this region (dark blue rectangles). Based on previous reports, there appear to be two distinct regions within Chromosome 16p11.2 associated with abnormal phenotypes, represented here by dashed boxes. The blue dashed box (“A”) corresponds to the locus most closely associated with BMI and the OMIM-associated genes deleted, while the gray dashed box (“B”) demarcates the locus that has been associated with seizure disorders, developmental delay, and autism spectrum disorders, along with the OMIM-associated genes deleted. In these three patients, the critical region is defined by Patient 1's deletion and corresponds with the BMI-associated locus. Patients 2 and 3, whose deletion includes both loci, also have neuro-developmental phenotypes, although neither of them are obese. None of the previously published patients whose deletion overlaps the critical region are reported to have renal developmental anomalies or Hirschsprung disease. Microdeletions based on the UCSC genome browser build 36/hg18, March 2006 assembly. A: BMI, CAKUT, and HSCR associated region. B: Autism/developmental delay associated locus. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com]
Patient 1 was found to have a de novo 217 kb deletion (minimal region: nucleotide 28,733,550–28,950,951, maximal region: 28,549,763–29,009,349) that overlapped with the larger deletion found in Patients 2 and 3. The smallest region of overlap between the three patients contained eight known genes (Table I).
| Gene | OMIM # | Disease associations | Function |
|---|---|---|---|
| ATXN2L | 607931 | Unknown | A novel component of the cytokine signaling system |
| TUFM | 610678 | Autosomal recessive disorder of combined oxidative phosphorylation deficiency presenting as lactic acidosis and encephalopathy | Human mitochondrial DNA nucleoid |
| SH2B1 | 608937 | Insulin resistance, glucose intolerance, hyperlipidemia, leptin resistance, hyperphagia, obesity | Mediates activation of various kinases. Functions in cytokine and growth factor receptor signaling and cellular transformation |
| ATP2A1 | 601003 | Autosomal recessive disorder of Brody myopathy | Sarcoplasmic reticulum Ca(2+) ATPase |
| RABEP2 | 611869 | Unknown | Endocytic membrane docking and fusion |
| CD19 | 107265 | Autosomal recessive disorder of antibody-deficiency | B-Lymphocyte antigen |
| SPNS1 | 612583 | Unknown | Transmembrane protein involved in necrotic cell death via autophagy |
| LAT | 602354 | Unknown | Regulates gamma-delta T cell homeostasis and differentiation |
DISCUSSION
We report on three boys with overlapping microdeletions within chromosome 16p11.2 who presented with a spectrum of CAKUT–HSCR anomalies. There are several regions within 16p11.2 that are predisposed to genomic alterations based on their architecture, including segmental duplications [Bailey et al., 2002; Sharp et al., 2006; Bochukova et al., 2010]. Several different regions within 16p11.2 have been associated with distinct phenotypes including aortic valve and seizure disorders [Ghebranious et al., 2007], developmental delays [Ballif et al., 2007; Bijlsma et al., 2009], autistic spectrum disorders [Kumar et al., 2008], and increased body mass index (BMI) [Bochukova et al., 2010] (Fig. 1).
The three boys reported here share a 217 kb deleted region that consists of eight genes (Table I). Based on the known biology of renal morphogenesis and enteric nervous system development, SH2B1 emerges as the most likely candidate gene of the eight.
Review of the literature showed several patients with 16p11.2 deletions that included SH2B1 (Fig. 1). Ballif et al. 2007 reported one patient with developmental delay whose deletion overlapped with the shortest region of overlap identified in the patients reported here. No renal or gastroenterologic abnormalities were noted in this patient. Bochukova et al. 2010 identified five patients with 16p11.2 deletions that included SH2B1 who presented with severe obesity. They did not report CAKUT–HSCR anomalies; however there was limited clinical data presented and systematic ultrasonography was not reported [Bochukova et al., 2010]. Of interest, neither of the two older patients reported here are obese.
SH2B1 is a member of the SH2B family of adaptor proteins, characterized by an SH2 domain, a pleckstrin homology domain, and a phenylalanine zipper dimerization domain [Donatello et al., 2007]. SH2B1 family members binds RET, nerve growth factor receptor (NGFR), insulin-like growth factor 1 receptor (IGF1R), insulin receptor (IR), janus kinase (JAK), platelet-derived growth factor (PDGFR), and fibroblast growth factor receptor (FGFR) [Maures et al., 2007]. SH2B1 has been shown to enhance the kinase activity of several receptor tyrosine kinases, including RET and has also demonstrated the ability to enhance neurite outgrowth [Zhang et al., 2006].
Using in vitro assays, Donatello et al. 2007 showed that SH2B1 enhances RET's tyrosine kinase activity. Co-expression of SH2B1 and RET was shown to increase autophosphorylation of RET and its downstream activation of SHC and STAT3. In addition, the binding of SH2B1 to RET at residues tyrosine 905 and tyrosine 981 protected RET from dephosphorylation by protein tyrosine phosphatases, resulting in prolongation of its activity. In vivo studies confirming that RET and SH2B1 are coexpressed in the enteric nervous system or developing kidney have yet to be published.
Through its involvement in mediating signaling between RET and GDNF, SH2B1 facilitates GDNF-induced neurite outgrowth in transfected cells and cultured mesencephalic neurons [Zhang et al., 2006; Chen et al., 2008]. Mutations in SH2B1 were associated with attenuated neurite outgrowth and decreased expression of important nerve growth factor (NGF) dependent genes [Chen et al., 2008]. Independent of the RET–GDNF pathway, SH2B1 has also been shown to bind NGF directly and enhance neurite outgrowth [Chen et al., 2008].
Our report of a new locus of interest on chromosome 16p11.2 associated with a CAKUT–HSCR phenotype provides a candidate region towards which studies can be conducted to help elucidate the common molecular pathogenesis underlying this association. Given the importance of the RET–GDNF pathway in renal branching morphogenesis and enteric enervation, it is compelling that the shortest region of overlap (SRO) in this deletion contains SH2B1, a gene potentially involved in RET signaling. Further laboratory investigation is needed to definitively determine whether loss of SH2B1 results in this phenotype, or whether one of the other genes in the SRO is related to the CAKUT–HSCR phenotype.




