Homozygous nonsense mutation in WNT10B and sporadic split-hand/foot malformation (SHFM) with autosomal recessive inheritance†
How to Cite this Article: Blattner A, Huber AR, Röthlisberger B. 2010. Homozygous nonsense mutation in WNT10B and sporadic split-hand/foot malformation (SHFM) with autosomal recessive inheritance. Am J Med Genet Part A 152A:2053–2056.
Abstract
Split-hand/foot malformation (SHFM) is a limb malformation affecting the central rays of the hands and/or feet. Isolated SHFM occurs within families but more often sporadically. Since most families with more than one patient show dominant inheritance with reduced penetrance, sporadic SHFM is generally considered to be due to dominantly inherited new mutations. Recently, recessive inheritance of SHFM was proposed in a highly consanguineous family with a homozygous missense mutation in WNT10B. Nevertheless, the assumption of a second locus was necessary to explain the observed phenotypes in this family. To date, no other family and no case of sporadic SHFM with WNT10B mutations are known. By examining WNT10B in a patient with sporadic SHFM, we identified a homozygous 4-bp duplication resulting in a premature termination codon. Nine heterozygous relatives show no sign of SHFM. These findings have profound implications for genetic counseling. Obviously, sporadic SHFM may show recessive rather than dominant inheritance resulting in a 25% recurrence risk for sibs instead of a very low-recurrence risk as generally presumed. Likewise, there is a very low-recurrence risk for offspring of patients (unless there is consanguinity) instead of an estimated risk between 30% and 50%. It can be concluded that sporadic SHFM is not always a dominant trait. To determine the recurrence risk, patients affected with sporadic SHFM should be tested for mutations in WNT10B. © 2010 Wiley-Liss, Inc.
INTRODUCTION
Split-hand/foot malformation (SHFM) is a congenital limb malformation involving the central rays of the hands and/or feet (OMIM SHFM1-6: 183600, 313350, 600095, 605289, 606708, and 225300). The observed phenotypes of SHFM are diverse, including ectrodactyly, syndactyly, and monodactyly. SHFM occurs either sporadically or within families. SHFM shows genetic heterogeneity and several loci have been mapped. SHFM1 (7q21), SHFM3 (10q24), SHFM4 (3q27), and SHFM5 (2q31) are dominantly inherited. Incomplete penetrance and variable expressivity have been documented in many pedigrees. Besides inter- and intra-familial variability, there are even differences among the limbs of a single individual. Due to the reduced penetrance, we estimated the recurrence risk for the next generation to be between 30% and 50%. This estimation is in agreement with the textbook of Harper 2004. According to this standard on genetic counseling, affected individuals have a high risk of having affected children even if the family pattern does appear to be autosomal recessive. In addition to these autosomal dominant inherited forms of SHFM, rare forms of SHFM have been described as X-chromosome linked (SHFM2, Xq26) or inherited as an autosomal recessive trait (SHFM6, 12q13). Recently, the study of a Turkish consanguineous family affected by SHFM6 led to the identification of a missense mutation in WNT10B exon 5 (R332W; WINGLESS-TYPE MMTV INTEGRATION SITE FAMILY, MEMBER 10B) [Ugur and Tolun, 2008]. The authors proposed recessive inheritance although the assumption of a second locus was necessary to explain the segregation of the phenotypes within this family. We report on the first sporadic patient with isolated SHFM with autosomal recessive inheritance due to a homozygous nonsense mutation in WNT10B. These findings have profound implications for genetic counseling of all patients with sporadic SHFM.
METHODS
Patient
The patient of this study was a pregnant 30-year-old Swiss woman with isolated sporadic SHFM, who was referred for genetic counseling and determination of the recurrence risk of SHFM for her offspring. SHFM was diagnosed at birth but detailed documentation is no more available. At 12 years, based on radiographs, the malformation was described as presenting with loss of second, third, and fourth toes of both feet, bifid fifth toe on the right foot and fusion of the third and fourth metatarsal of both feet. A surgical osteotomy of the fourth and fifth metatarsal on the right foot was performed at 5 years. The right hand impressed at birth with complete cutaneous syndactyly of the fourth and fifth finger, hypoplasia of the third finger with loss of the distal phalanx. The right hand showed proximal cutaneous syndactyly of the third and fourth finger. Syndactyly was surgically corrected at 2 years. At the time of counseling, radiographs were no longer available. No mutation was detected previously in TP63 (TUMOR PROTEIN p63) by direct sequencing (data not shown). Consanguinity of the parents of the patient is not known. A detailed pedigree is shown in Figure 1.
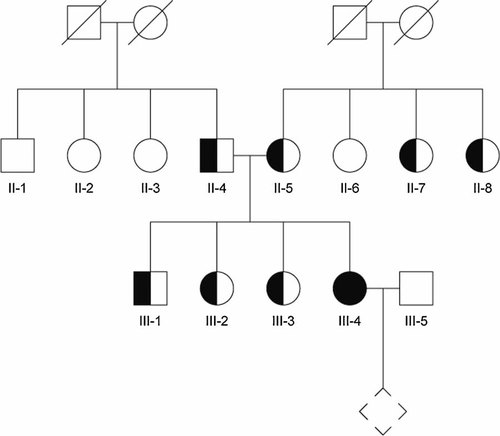
Pedigree with sporadic split-hand/foot malformation (SHFM). The patient with a homozygous WNT10B mutation (c.458_461dupAGCA) and isolated SHFM is indicated in black. Molecular diagnosis was performed in all numbered individuals (II-1, II-2, etc.). Confirmed heterozygotes are indicated as half-black, half-white symbols.
Sequencing of WNT10B
DNA was isolated from peripheral blood lymphocytes using the MagNA pure compact device (Roche, Rotkreuz, Switzerland). Primers and PCR conditions for WNT10B amplification are available on request. The purified PCR-products were sequenced in both directions and visualized on an ABI 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA).
Real-Time Quantitative WNT10B RT-PCR
Total RNA was extracted using the PAXgene blood RNA kit according to the manufacturer's protocol (Qiagen, Hilden, Germany). Reverse Transcription was done using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). For real-time PCR, a commercially available TaqMan® gene expression assay specific for WNT10B was used (Hs00928823_m1; Applied Biosystems) and the absolute quantification of ABL RNA served as endogenous control (Ipsogen SA, Marseille, France). Real-time PCR was carried out in a reaction volume of 20 µl and 50 amplification cycles with 10 sec at 95°C and 1 min at 60°C. Amplification and analysis were performed on a LightCycler® 2.0 (Roche).
RESULTS
Sequencing of the entire WNT10B coding region documented a 4-bp homozygous duplication in exon 4 (c.458_461dupAGCA, GenBank accession no. NM_003394). The duplication of AGCA leads to a frameshift and results in a premature TAA termination codon after 46 altered amino acids (p. Asp 155AlafsX47). The described duplication was found in both parents of the patient in a heterozygous form. Furthermore, WNT10B exon 4 was sequenced in all three sibs of the patient and in all six parental sibs. The brother and both sisters were heterozygous carriers of the duplication. Among the sibs of the parents four were carriers of the wild-type allele and two were duplication heterozygotes (Figs. 1 and 2). Neither the heterozygous carriers nor the individuals harboring only the wild-type allele showed any signs of SHFM or any other limb malformation. Sequencing of the entire WNT10B coding region in the partner of the patient revealed no mutation.
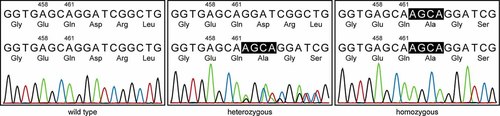
Sequencing analysis of WNT10B. Electropherograms of the 4 bp-duplication (c.458_461dupAGCA) are shown for all three genotypes (wild type, heterozygous, and homozygous). The base pair sequences and the amino acid translations are shown for both alleles, respectively. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
For the relative quantification of WNT10B levels, a standard curve using five standard dilutions was constructed (data not shown). For each sample, the amount of WNT10B RNA was determined with the help of the corresponding standard curve. To obtain normalized values, the relative amount of WNT10B RNA was divided by the absolute amount of ABL endogenous control RNA (WNT10B/ABL). The WNT10B/ABL value as a measure of WNT10B gene expression level was 3.90E−4 ± 1.89E−4 for the four wild type individuals and 5.15E−4 ± 1.63E−4 for the seven duplication heterozygotes. The WNT10B/ABL value of the patient was 3.62E−4 and did not significantly differ from the values above (t-test).
DISCUSSION
Only few reports of SHFM are consistent with X-linked inheritance (SHFM2, Xq26) [Faiyaz-Ul-Haque et al., 2005] or autosomal recessive inheritance (SHFM6) [Klein, 1932; Ray, 1970; Freire-Maia, 1971; Verma et al., 1976; Zlotogora and Nubani, 1989; Gul and Oktenli, 2002]. Therefore, sporadic patients of isolated SHFM are considered to be generally the result of de novo dominant mutations. As a consequence, the recurrence risk for sibs of patients with sporadic SHFM is estimated to be very low while the recurrence risk for offspring of patients is estimated to be between 30% and 50% [Harper, 2004]. Recently, the study of a large consanguineous family with isolated SHFM6 led to the identification of a missense mutation in exon 5 (R332W) in WNT10B on chromosome 12 [Ugur and Tolun, 2008]. This mutation was present in a homozygous state in all affected individuals except for one. In addition, there was one individual without any developmental defect of the limbs despite harboring the homozygous mutation. The authors propose a model suggesting either the contribution of a second locus or the presence of a suppressor mutation in the nonpenetrant individual. To our knowledge, in addition to the family described by Ugur and Tolun 2008, in no other family with isolated SHFM and in no sporadic patient of SHFM, mutations in WNT10B have been described to date. The patient described in our study was a Swiss woman with sporadic and isolated SHFM. At the time of molecular investigation she was pregnant. The research aspect of the molecular investigation was explicitly discussed with the patient and she was informed about the possibility that mutations with unknown significance could be detected (e.g., missense mutations). Anyway, the patient did not consider an abortion, irrespective of the molecular result. The 4-bp homozygous duplication found in this patient in exon 4 of the WNT10B gene (c.458_461dupAGCA) results in a premature TAA termination codon after 46 altered amino acids. Also, 11 unaffected relatives were tested for the presence of the mutation in exon 4 of WNT10B and all carried either none or only one deleterious allele. Measurements of WNT10B transcript levels by real-time PCR showed no difference between the carriers of homozygous, heterozygous, and wild-type alleles (data not shown). These findings strongly suggest autosomal recessive inheritance, which has profound implications for genetic counseling. Obviously, sporadic SHFM may show recessive rather than dominant inheritance which results in a 25% recurrence risk for sibs instead of very low-recurrence risk as generally presumed; and in a very low-recurrence risk for offspring of affected patients instead of a risk between 30% and 50% (as long as there is no consanguinity). As mentioned above, sequencing of the entire WNT10B coding region in the partner of the patient revealed no mutation. We conclude that sporadic SHFM is not always of the dominant type and, to determine the risk of recurrence, patients affected by sporadic SHFM should be tested for mutations in WNT10B.
Acknowledgements
We would like to thank the family that participated in the study as well as our laboratory technicians for their skillful help.




