Myoclonus dystonia plus syndrome due to a novel 7q21 microdeletion†
How to Cite this Article: Saugier-Veber P, Doummar D, Barthez M-A, Czernecki V, Drouot N, Apartis E, Bürglen L, Frebourg T, Roze E. 2010. Myoclonus dystonia plus syndrome due to a novel 7q21 microdeletion. Am J Med Genet Part A 152A:1244–1249.
Abstract
Myoclonus dystonia (M-D) is a rare genetic movement disorder characterized by a combination of myoclonic jerks and dystonia. It is usually due to mutations in the SGCE gene. We report on a patient with a typical M-D syndrome, but also short stature, microcephaly, and mental retardation. Molecular analysis showed no mutations within the SGCE gene but a microdeletion encompassing the SGCE gene in chromosome region 7q21. Array-CGH analysis showed that the deletion spanned approximately 1.88 Mb. We suggest that M-D plus patients with mental retardation, microcephaly, dysmorphism, or short stature, all frequently associated disorders, should be screened for 7q21 microdeletion. By examining previously published cases of mental retardation associated with pure 7q21 deletions, we identified two distinct regions of respectively 455 and 496 kb that are critical for mental retardation and growth retardation. Among the genes located within these regions, LOC253012, also known as HEPACAM2, is a good candidate for both mental retardation and microcephaly. © 2010 Wiley-Liss, Inc.
INTRODUCTION
Myoclonus dystonia (M-D) is a rare movement disorder characterized by a combination of myoclonic jerks and dystonia (DYT11; OMIM#159900). Clinical manifestations generally occur in the first or second decade of life, and myoclonus, predominating in the arms and axial muscles, is usually the main and most disabling feature [Roze et al., 2008]. M-D is an autosomal dominant disorder that can be caused by mutations in the epsilon-sarcoglycan (SGCE) gene, located in chromosome region 7q21 [Zimprich et al., 2001; Kinugawa et al., 2009]. Penetrance is strongly dependent on the parental origin of the disease allele, maternal imprinting being associated with lower penetrance of the phenotype upon maternal inheritance of the mutated allele [Zimprich et al., 2001]. Mutations or large deletions of the SGCE gene are detected in about 40% of patients with the typical phenotype, suggesting that the disorder is genetically heterogeneous [Tezenas du Montcel et al., 2006]. Mutations or intragenic deletions within the SGCE gene account for almost all cases of M-D syndrome due to SGCE deficiency. In the remaining SGCE-deficient patients, large chromosomal microdeletions encompassing the entire SGCE gene of paternal origin, and adjacent genes, can result in M-D associated with additional clinical features: the so-called “M-D plus syndrome” [Asmus et al., 2007; Bonnet et al., 2008]. We report on a new case of M-D due to a microdeletion in chromosome region 7q21 and discuss the need to take into account associated clinical features when designing molecular analyses.
PATIENTS AND METHODS
Clinical Report
The patient, a 17-year-old man, developed a movement disorder at the age of 12 years. Thereafter, his condition gradually worsened. The pregnancy and delivery were normal, except for unexplained intrauterine growth retardation. At birth, at 39 weeks of gestation, he had a weight of 2,420 g (−2 SD), a height of 46.5 cm (−2 SD), and an occipitofrontal circumference of 32.5 cm (−1.5 SD). During childhood and adolescence, he had growth retardation and delayed psychomotor development and was always noted to be clumsy with poor coordination. When referred to us at age 17 years, he measured 151 cm (−3.5 SD), had a weight of 35 kg (−4 SD) and an occipitofrontal circumference of 51 cm (−3 SD). On examination, he had shock-like, mainly proximal, asymmetrical and asynchronous myoclonic jerks of the four limbs and trunk, that predominated on the upper body. He also had mild generalized dystonia that also predominated on the upper body. Neuropsychological examination showed mental retardation with an IQ of 30. He had mild hyperlaxity of the fingers and ankles but no other features of osteogenesis imperfecta. Skeletal radiography showed scoliosis at the lower thoracic level but no bone dysplasia or joint abnormalities. Brain MRI was normal, and neurophysiological studies showed sub-cortical myoclonus. His father, seen at age 50 years, has a milder but similar phenotype with asymmetrical asynchronous myoclonic tremor of the upper limbs, mild segmental dystonia involving the neck and right upper arm, mild mental retardation, microcephaly (52.5 cm), and short stature (156 cm).
Sequencing Analysis
Blood samples were collected with the patients' written informed consent.
The SGCE gene is composed of 12 exons. Exon 10 is alternatively spliced and is missing in the majority of transcripts. The 11 exons and intron–exon junctions of the major SGCE transcript (NM_003919.2) were PCR-amplified using 11 primer pairs (sequences available on request). The PCR products were then sequenced with Big Dye Terminator cycle sequencing kit on an ABI Prism 3130xl automated sequencer (PE Applied Biosystems, Foster City, CA).
Quantitative Multiplex PCR of Short Fluorescent Fragments (QMPSF)
Short fragments (140–260 bp) of the 11 exons encoding the major SGCE transcript (NM_003919.2) were simultaneously amplified by multiplex PCR with dye-labeled primers corresponding to unique sequences (see supporting information Table I which may be found in the online version of this article). Two additional fragments, corresponding to exon 13 of the HMBS gene located on chromosome 11, and exon 3 of the MECP2 gene located on chromosome X, were co-amplified as controls. PCR was performed in quantitative conditions as previously described [Charbonnier et al., 2000]. DNA fragments generated by QMPSF were separated on an Applied Biosystems model 3130xl automated sequencer (PE Applied Biosystems). The data were analyzed with Gene Mapper software version 4.0 (PE Applied Biosystems). The PCR kinetics was controlled by superimposing the electropherograms of a normal male control DNA and a normal female control DNA, the height of the MECP2 peak being 50% lower in the male control DNA. The heights of the SGCE exon peaks were then compared between the patients and normal male and female controls.
Molecular Karyotyping
In order to determine the extend of the deletion, we performed array-CGH (Agilent, Agilent Technologies, Santa Clara, CA) using 105,000 oligo probes spaced at intervals of approximately 20 kb across the genome (Human Genome CGH microarray 105B kit, Agilent). Commercial male genomic DNA (Promega, Madison, WI) was used as reference. Hybridization results were extracted with feature extraction software and analyzed with DNA analytic software by applying an ADM 2 segmentation algorithm to identify chromosome aberrations.
RESULTS
Sequencing analysis of the 11 exons encoding the major SGCE transcript (NM_003919.2) revealed no mutations. We then used QMPSF to screen for deletions and duplications and thus to ensure accurate molecular diagnosis of the M-D syndrome. We identified a partial deletion of the gene, removing exons 2–11 but respecting exon 1 (Fig. 1A), showing that the distal breakpoint is located in intron 1. Using array-CGH to determine the extent of the deletion, we showed that the proximal breakpoint was located in intron 3 of the CDK6 gene (NM_001259.5) (Fig. 1B), giving an estimated deletion size of 1.88 Mb (arr 7q21(92,241,997–94,120,153)×1 pat). The boy's father shared the same chromosomal defect (data not shown).
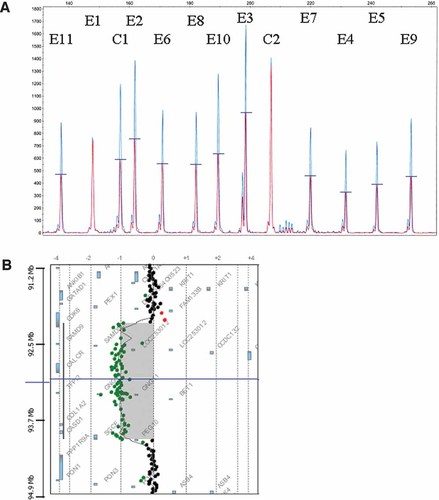
QMPSF detection of a 7q21 microdeletion. A: QMPSF exploring the 11 exons encoding the major SGCE transcript (NM_003919.2) revealed that the patient harbors a deletion removing 10 of the 11 exons. The patient's electropherogram (red) was superimposed on that of a normal female control (blue) by adjusting to the same level the peaks obtained for the control amplicon (C2) located in the HMBS gene on 11q23. The other control amplicon (C1), located in the MECP2 gene on the X chromosome, controls the amplification kinetics (see supporting information Table I which may be found in the online version of this article). The numbers above the peaks indicate the SGCE exons. The Y-axis displays fluorescence in arbitrary units, and the X-axis indicates the size in bp. Heterozygous deletions are evident from the 50% lower peaks in the patient compared to the normal control. B: Array-CGH profiling of chromosome 7 using the Agilent 105K microarray (DNA analytics software display) showing the 7q21 microdeletion. The proximal breakpoint of the deletion is located between 92,220,116 and 92,241,997 while the distal breakpoint is located between 94,120,153 and 94,134,765 according to the Human Gene Assembly hg18. Based on the results of the QMPSF assay, we refined the location of the distal breakpoint within the 3-kb interval to 94,120,153–94,123,237.
DISCUSSION
This patient had the typical clinical features of M-D syndrome, but also short stature, microcephaly, and mental retardation. Molecular analysis showed no mutations within the SGCE gene but identified a microdeletion encompassing the SGCE gene in the 7q21 chromosome region. Such cases of M-D with complex phenotypes may be under diagnosed, as additional features are classically exclusion criteria for the M-D syndrome [Kinugawa et al., 2009]. We therefore insist that patients with M-D as well as mental retardation, microcephaly, dysmorphism or short stature should be screened for 7q21 microdeletion. This is an important issue, as methods for detecting mutations and deletions are different. Although he shares the same genetic defect, the propositus' father has a similar but milder phenotype. Intrafamilial phenotypic variability has already been described in families with classical M-D syndrome due to SGCE deficiency [Kinugawa et al., 2009] and in other forms of dominant primary dystonia, including dystonia due to torsin A deficiency or GTP cyclohydrolase 1 deficiency [Robison et al., 1999; Opal et al., 2002]. Such intrafamilial variability suggests the influence of genetic or epigenetic modifying factors on disease progression.
Microdeletions involving the SGCE gene have been described as a cause of the M-D syndrome [DeBerardinis et al., 2003; Asmus et al., 2005, 2007; Bonnet et al., 2008; Grünewald et al., 2008; Han et al., 2008]. Two types of deletion can be distinguished: (i) partial deletions of the SGCE gene corresponding to exonic deletions and resulting in the typical M-D syndrome; and (ii) large deletions encompassing the flanking genes, resulting in complex phenotypes including M-D. At the present time, seven reports of such large 7q21 deletions involving the SGCE gene have been reported [DeBerardinis et al., 2003; Asmus et al., 2007; Bonnet et al., 2008; Grünewald et al., 2008]. It is worth noting that no recurrent breakpoint has so far been found. Therefore, these large deletions may arise either by nonhomologous recombination with homologous end-joining or by fork stalling and template switching [Lee et al., 2007].
Symptoms associated with M-D in this setting include cerebral cavernous malformations, mild bone and joint disorders, split hand/split foot malformation, hearing loss, short stature, microcephaly, mental retardation, and facial dysmorphism. Cerebral cavernous malformations are due to deletion of the KRIT1 gene, bone and joint disorders to deletion of the COL1A2 gene, split hand/split foot malformations to deletion of the SHFM1 gene, and sensorineural hearing loss to deletion of the DLX6-DLX5 genes. The genes involved in short stature, mental retardation, microcephaly, and facial dysmorphism have yet to be identified. Here, the detection of a SGCE deletion involving exons 2–11 in a patient with a complex phenotype characterized by M-D and short stature, microcephaly, and mental retardation led us to perform fine mapping of the rearrangement. We found that this deletion spanned a 1.88-Mb genomic region corresponding to one of the smallest reported 7q21 deletions. KRIT1, SHFM, and DLX6-DLX5 were not deleted. The COL1A2 gene was deleted but the patient had no manifestations of osteogenesis imperfecta, except for finger and ankle hyperlaxity and scoliosis, although the latter could have been due to long-standing axial dystonia. In addition, array-CGH studies allowed us to exclude another associated chromosomal anomaly as the cause of short stature and mental retardation.
Deletion of the 7q21 region is frequently associated with mental retardation and short stature. In view of previous observations of a Silver-Russell patient with uniparental (maternal) disomy of chromosome 7 and no mental retardation, we infer that distinct genes are responsible for short stature and mental retardation [Guettard et al., 2008]. In our patient the deleted genomic segment encompasses 19 genes, none of which have previously been implicated in mental retardation or growth retardation. By analyzing previously published cases of mental retardation associated with pure 7q21 deletion [Scherer et al., 1994; DeBerardinis et al., 2003; Fukushima et al., 2003; Wieland et al., 2004; Courtens et al., 2005; Asmus et al., 2007; Tzschach et al., 2007] and, more precisely, by comparing the 1.63-Mb deletion in Patient #1 of Asmus et al. 2007 and the 1.88-Mb deletion in our patient, we identified a critical 455-kb region (92,241–92,696K) including only six genes, namely CDK6, LOC100128994, RN7SLP4, SAMD9, SAMD9L, and LOC253012/HEPACAM2 (Fig. 2). CDK6 and LOC253012/HEPACAM2 (recently named Miki for mitotic kinetics regulator), both of which are involved in the regulation of cell proliferation, are candidate genes for mental retardation. Miki has been shown to localize to centrosomes/spindles during mitosis [Asou et al., 2009]. Interestingly, five genes encoding centrosomal proteins have been implicated in primary microcephaly, which is always associated with mild to severe mental retardation [Kumar et al., 2009]. LOC253012/HEPACAM2 could thus be a good candidate gene for mental retardation and microcephaly. Alternatively, this 455-kb region could contain a regulatory element involved in long-range control of gene expression [Kleinjan and van Heyningen, 2005]. However, another patient described by Asmus et al. 2007 (Patient #2), who had a deletion encompassing the critical region, had no reported mental retardation. The reason for this is unclear. Comprehensive analysis of pure 7q21 deletions in patients with and without short stature [Scherer et al., 1994; DeBerardinis et al., 2003; Fukushima et al., 2003; Wieland et al., 2004; Courtens et al., 2005; Asmus et al., 2007; Tzschach et al., 2007; Grünewald et al., 2008] allowed us to identify a distinct critical region of 496 kb (92,696–93,192K), which contains five genes, namely CCDC132, MIRN653, MIRN489, CALCR, and LOC392748 (Fig. 2). CALCR, encoding the calcitonin receptor, that plays a role in bone mineralization, is a possible candidate for growth retardation. Two miRNA genes, MIRN653 and MIRN489, are other possible candidates. MicroRNAs (miRNAs) are small, noncoding RNAs which bind to complementary sites on the 3′ UTR of dozens of target mRNAs to modulate their expression, most often by inhibiting protein translation. They are important in the regulation of cell proliferation and differentiation, organogenesis, and progenitor cell maintenance [Kloosterman and Plasterk, 2006]. Fine mapping of novel 7q21 deletions should help to identify new genes involved in mental retardation, microcephaly, and short stature.
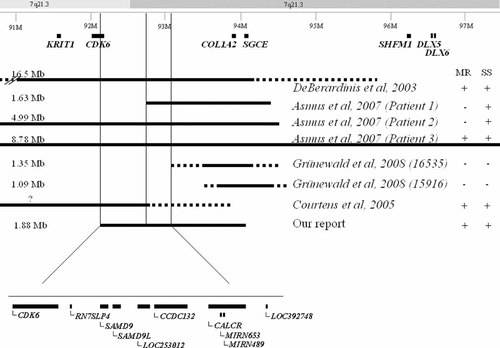
Schematic representation of chromosome 7, indicating the deleted region in our patient and other reported large 7q21 deletions. The presence (+) or absence (−) of mental retardation (MR) and of short stature (SS) is indicated on the right. The exact gene content of the 1-Mb region critical for MR and SS is described at the bottom of the figure.
ELECTRONIC DATABASE INFORMATION
Accession numbers and URLs for data presented herein are as follows:
GenBank, http://ncbi.nih.gov/GenBank/ (for SGCE cDNA, accession number NM_003919.2); Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/.
Acknowledgements
We thank Constance Flamand-Rouvière for helping to prepare the manuscript.




