Interstitial deletion 2p11.2–p12: Report of a patient with mental retardation and review of the literature†
How to cite this article: Tzschach A, Graul-Neumann LM, Konrat K, Richter R, Ebert G, Ullmann R, Neitzel H. 2009. Interstitial deletion 2p11.2–p12: Report of a patient with mental retardation and review of the literature. Am J Med Genet Part A 149A:242–245.
Abstract
Deletions of chromosome bands 2p11.2 and 2p12 are rare, and only six patients have been reported to date. Here, we report on a 5-year-old girl with an 11.4 Mb interstitial deletion of chromosome bands 2p11.2–p12 and the characterization of this deletion by high-resolution array CGH. The patient presented with mental retardation, microcephaly and short stature. Facial features included broad nasal bridge, frontal bossing and mild dolichocephaly. Phenotypic comparison with previously published patients failed to reveal a consistent clinical pattern apart from developmental delay/mental retardation, which is probably due to different sizes and/or positions of the individual deletions. Among the 40 known genes deleted in our patient is REEP1, haploinsufficiency of which causes autosomal dominant spastic paraplegia type 31 (SPG31, OMIM 610250). Additional patients with well-characterized deletions within 2p11.2 and 2p12 will be needed to determine the role of individual genes for the clinical manifestations. © 2009 Wiley-Liss, Inc.
INTRODUCTION
Interstitial deletions of chromosome bands 2p11.2–p12 are rare, and only six patients have been reported to date [Prasher et al., 1993; Los et al., 1994; Wenger and McPherson, 1997; Lacbawan et al., 1999; Barber et al., 2005]. These patients had developmental delay/mental retardation and variable additional problems such as hypospadias, facial dysmorphisms, club foot or Wilms tumor (Table I).
|
Clinical manifestation |
Prasher et al. 1993 |
Los et al. 1994 |
Wenger and McPherson 1997 |
Lacbawan et al. 1999 |
Barber et al. 2005 , Family 3, son |
Barber et al. 2005 , Family 3, mother |
Present patient |
|---|---|---|---|---|---|---|---|
| Position of deletion in 2p | p11.2–p13 | p11.2–p13 | p11.2–p13 | p11.2–p13 | p11.2–p12 | p11.2–p12 | p11.2–p12 |
| Size of deletion | NA | NA | NA | NA | 7.5 Mb | 7.5 Mb | 11.4 Mb |
| Sex | m | m | m | m | m | f | f |
| Age of examination | 32 years | 4 years | 2 months (died) | 1 year | 4 years 9 months | Adult | 5 years |
| Developmental delay | + | + | Died at age 2 months | + | + | “Limited intellectual resources” | + |
| Growth retardation | − | + | 90th centile at birth | 75th centile at birth | NA | NA | + |
| Head circumference | 53 cm (<25th cent.) | 10th centile | 10th centile at birth | <3rd centile at birth | Large head | NA | 47.5 cm (<3rd centile) |
| Facial anomalies | Blepharophimosis, broad nasal bridge | Short palpebral fissures, ptosis | Pointed nose | Epicanthal folds, broad nasal bridge | Flat face, low-set abnormally molded ears | Abnormally molded ears | Broad nasal bridge, mild dolichocephaly |
| Abnormal ears | + | + | + | Simple/cupped ears | + | + | − |
| Frontal bossing | − | + | − | − | + | NA | + |
| Genital malformation | − | Cryptorchidism, hypospadias | Cryptorchidism | − | − | − | − |
| Abnormalities of chest or spine | Narrow chest | − | Scoliosis, pectus carinatum | − | − | − | Pectus carinatum, hyperlordosis |
| Digital contractures | Distal interphalangeal joints of fingers | Clasped thumb | + | − | − | − | − |
| Other problems | Thrombocytopenia, hypertonia, spastic ataxic gait | Clubfoot, vitiligo, hearing impairment | Dextrocardia, VSD, eventration of diaphragm | Deafness, cyst at left neck | Wilms tumor | − | Clubfoot, single umbilical artery |
Here, we report on the characterization of a 2p11.2–p12 deletion by tiling-path array comparative genomic hybridization (array CGH) in a mentally retarded female patient, and we compare her clinical manifestations with the previously published patients.
CLINICAL REPORT
The girl was the fourth child of healthy and nonconsanguineous parents; she has three healthy brothers. Ultrasound examination during pregnancy revealed bilateral clubfoot and a single umbilical artery. She was born at term by normal delivery [birth weight 3,230 g (10th–25th centile); length 50 cm (10th–25th centile); head circumference 34 cm (10th centile); APGAR 10/10]. Clubfeet were surgically corrected at the age of 6 months and again at the age of 4 years. Psychomotor development was retarded; she spoke first words at the age of 2 years and started walking at 3 years.
On examination at the age of 5 years, she was short and mildly microcephalic [head circumference 47.5 cm (<3rd centile), height 102 cm (3rd centile), weight 11.8 kg (<3rd centile)]. Physical examination revealed mild cutaneous syndactylies on hands and feet, pectus carinatum and hyperlordosis. Facial dysmorphic signs included low-set ears, broad nasal bridge, frontal bossing and mild dolichocephaly (Fig. 1). A brain MRI scan at the age of 3 years revealed no abnormalities. She was a friendly girl with a cheerful disposition.
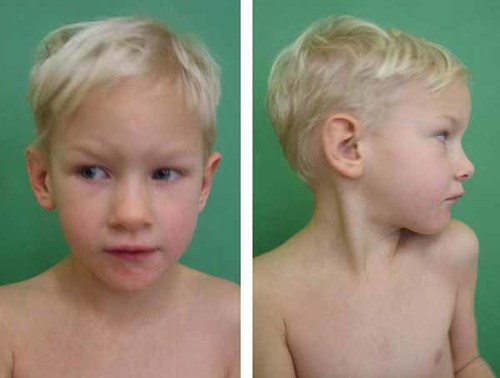
Facial appearance of the patient at the age of 5 years. Note mild dolichocephaly, broad nasal bridge and prominent forehead. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
MATERIALS AND METHODS
Karyotyping
Cytogenetic investigations (GTG banding) on 25 metaphases obtained from PHA-stimulated peripheral lymphocytes were performed according to standard protocols after informed consent.
Array CGH
Hybridizations were performed on a tiling path BAC array as described previously [Erdogan et al., 2006]. Aberrations were only considered if at least three adjacent clones were involved unless they coincided with published DNA copy number variants as listed in the Database of Genomic Variants (http://projects.tcag.ca/variation/). For estimating the content of segmental duplications (low copy repeats), each BAC clone was classified into one out of seven categories and colored as described previously [Chen et al., 2005]. Detailed step-by-step protocols are provided on our website (http://www.molgen.mpg.de/∼abt_rop/molecular_cytogenetics/).
RESULTS
Karyotyping
Chromosome analysis in the patient revealed an interstitial deletion of the proximal short arm of one chromosome 2 in all metaphases [karyotype 46,XX,del(2)(pter->p13::p11.2->qter)] (Fig. 2a). Both parents had normal karyotypes.
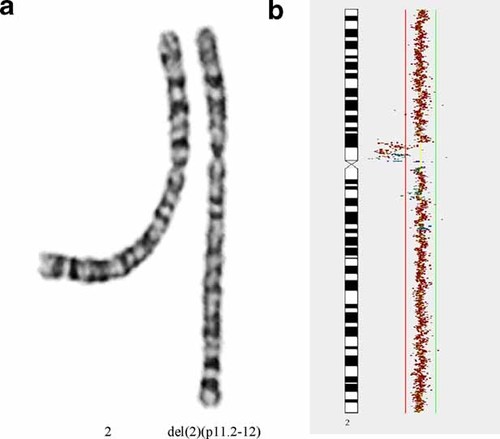
a: Partial karyogram (GTG banding) showing the normal and the deleted chromosome 2. b: Partial array CGH results as displayed by CGHPro for the deleted region of chromosome 2. The ratios of the clones are plotted in a size dependent manner along the chromosome ideograms. The green and red lines represent log2 ratios of 0.3 (gains) and −0.3 (losses), respectively. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Array CGH
Whole genome array CGH confirmed the deletion in 2p. The distal breakpoint of the deletion was defined by the overlapping BAC clones RP11-768O23 (last normal clone, position 79,853,483–80,031,025) and RP11-146A20 (first deleted clone, position 79,960,926–80,140,468). The proximal breakpoint was mapped to BAC clones RP11-458O20 (last deleted clone, position 91,151,313–91,362,960) and RP11-205B04 (first normal clone, position 91,298,566–91,448,461) immediately adjacent to the centromeric region (all genomic positions are given according to UCSC genome browser May 2004 assembly) (Fig. 2b). Thus, the deletion had a size of approximately 11.4 Mb. The karyotype according to the ISCN 2005 nomenclature is 46,XX.arr cgh 2p11.2p12(RP11-146A20–>RP11-458O20) × 1.
DISCUSSION
We have listed the clinical manifestations of the six previously published patients with deletions in 2p11.2–p12 and of our patient in Table I [Prasher et al., 1993; Los et al., 1994; Wenger and McPherson, 1997; Lacbawan et al., 1999; Barber et al., 2005]. Apart from developmental delay/mental retardation, there were no other consistent findings which would allow the delineation of a clinically recognizable 2p11.2–p12 deletion syndrome. Phenotype-karyotype correlations are, however, restricted by the fact that most deletions in Table I had only been analyzed by GTG banding, and the precise positions and sizes of these deletions are therefore uncertain. Only the deletion reported by Barber et al. 2005 (family 3) had been characterized by microsatellite analysis. This deletion (7.5 Mb) is smaller on the proximal part of 2p11.2 than the deletion of our patient (11.4 Mb), which may provide an explanation why, in contrast to the patient reported here, their patients had neither short stature nor microcephaly.
While chromosome band 2p12 contains very few genes and deletions restricted to this band apparently cause no adverse effects [Barber et al., 2005], there are more than 40 known genes located in chromosome band 2p11.2. One of these genes is REEP1, haploinsufficiency of which causes autosomal dominant spastic paraplegia 31 (SPG31, OMIM 610250) [Zuchner et al., 2006]. It is noteworthy that the 32-year-old patient reported by Prasher et al. 1993 suffered from hypertonia of both legs and had a spastic, ataxic gait; these problems are suggestive of spastic paraplegia. The other patients with 2p11.2 deletions were probably too young to show clear signs of spastic paraplegia.
For the time being it must remain speculative which genes within the deleted interval play a major role for the clinical manifestations. Only the two most distal genes—CTNNA2 and LRRTM1—can probably be excluded as candidate genes because they had also been deleted in the healthy carriers of 2p12 deletions reported by Barber et al. 2005 (families 1 and 2).
On the other hand, only the 3′ part of the CTNNA2 gene is deleted in our patient, and a dominant negative effect caused by a C-terminal truncated CTNNA2 protein may also contribute to the phenotype.
In conclusion, the report of this patient adds to our knowledge of the clinical consequences of 2p11.2–p12 deletions. Additional patients with well-characterized deletions in this region will be needed for better karyotype–phenotype correlations and to dissect the role of individual genes for the clinical manifestations.
ELECTRONIC DATABASE INFORMATION
CGHPro, http://www.molgen.mpg.de/∼abt_rop/molecular_cytogenetics/ArrayCGH/CGHPRO.
Protocols for array CGH, http://www.molgen.mpg.de/∼abt_rop/molecular_cytogenetics/ProtocolsEntry.html.
Database of Genomic Variants, http://projects.tcag.ca/variation/.
Acknowledgements
We thank the parents of the patient for their support and Gabriele Scholz and Ines Müller for expert technical assistance. Array CGH was supported by the German Nationales Genomforschungsnetzwerk, grant 01GR0203. We would like to thank the BACPAC Resources Centre (http://bacpac.chori.org), the COST B19 Action “Molecular Cytogenetics of Solid tumors” and the Mapping Core and Map Finishing groups of the Wellcome Trust Sanger Institute for initial clone supply and verification.




