Molecular characterization of a patient with 3p deletion syndrome and a review of the literature†‡
How to cite this article: Fernandez TV, García-González IJ, Mason CE, Hernández-Zaragoza G, Ledezma-Rodríguez VC, Anguiano-Alvarez VM, E'Vega R, Gutiérrez-Angulo M, Maya ML, García-Bejarano HE, González-Cruz M, Barrios S, Atorga R, López-Cardona MG, Armendariz-Borunda J, State MW, Dávalos NO. 2008. Molecular characterization of a patient with 3p deletion syndrome and a review of the literature. Am J Med Genet Part A.
Thomas V. Fernandez and I.J. García-González contributed equally to this work.
Abstract
3p deletion syndrome is a rare disorder involving developmental delay, dysmorphic physical features, and growth retardation. Molecular mapping of several cases in the literature have identified a critical region on chromosome 3p26. We present a child patient with characteristic features of 3p deletion syndrome and a de novo unbalanced translocation involving chromosomes 3 and 13. Fine mapping of this rearrangement using fluorescence in situ hybridization (FISH) and array-based comparative genomic hybridization (aCGH) revealed an unbalanced abnormality including a 4.5 Mb terminal deletion of chromosome 3p, telomeric to ITPR1 on 3p26.2, which was not previously identified with routine cytogenetic analysis. In addition, these investigations confirmed and refined the boundaries of a 26.5 Mb deletion of chromosome 13. This study confirms the minimal candidate region for 3p deletion syndrome, provides further evidence implicating haploinsufficiency of CNTN4 in the disorder, and demonstrates the utility of high-resolution investigations of rare chromosomal rearrangements. © 2008 Wiley-Liss, Inc.
INTRODUCTION
3p deletion syndrome, first described by Verjaal and De Nef 1978, is characterized by developmental delay, dysmorphic facial features and growth retardation [Verjaal and De Nef, 1978; Angeloni et al., 1999; Sotgia et al., 1999; Kariya et al., 2000; Cargile et al., 2002; Fernandez et al., 2004]. Since then, a number of affected individuals with circumscribed, overlapping regions of contiguous haploinsufficiency on chromosome 3p have been identified, defining a 1.5 Mb critical region on chromosome 3p26 [Cargile et al., 2002; Dijkhuizen et al., 2006]. A recent report of a de novo balanced translocation in a boy with this syndrome identified the disruption of a single copy of Contactin 4 (CNTN4), a brain-expressed gene within this critical region on chromosome 3p26.3, as sufficient to confer key aspects of the phenotype [Fernandez et al., 2004].
We present a child patient with the characteristic physical features of 3p deletion syndrome, carrying a de novo translocation involving chromosomes 3p and 13q that was further characterized using fluorescence in situ hybridization (FISH) and array-based comparative genomic hybridization (aCGH). These molecular studies demonstrate that the propositus possesses an unbalanced chromosomal abnormality including a deletion of chromosome 3p telomeric to ITPR1. The study confirms the minimal candidate region for 3p- syndrome and demonstrates the value of array-based cytogenetic evaluation in such instances.
CLINICAL REPORT
The propositus is a 5½-year-old male who was the product of the 2nd full normal pregnancy of young, healthy, and nonconsanguineous parents. Birth weight was 3.0 kg (10th centile) and height was 50.5 cm (50th centile). A cleft lip and palate was detected at birth and psychomotor development was delayed.
Physical exam of the patient was undertaken after appropriate consents, approved by our institutional review board, were obtained. He was noted to have a weight of 16.5 kg (<3rd centile), a height of 102.5 cm (<3rd centile), and occipital frontal circumference (OFC) of 48 cm (<3rd centile). Language delay was seen. Notable physical features included plagiocephaly, microcephaly, triangular face, flat and narrow fontanelle, sparse eyebrows with inverted “V” form, downslanting palpebral fissures, broad and high nasal bridge, hypoplastic nose, bifid uvula, and absent philtrum. Ears were dysplastic, low-set and posteriorly rotated with an unraveled right ear with a preauricular tag. The mouth was small with downturned corners, thin lips, a wide-high palate with a surgically repaired posterior cleft palate and lip, and a bifid uvula. He also presented with retromicrognathia, short neck and tight thorax (Fig. 1, Table I). The hands showed brachydactyly and cutaneous syndactyly in all fingers and bilateral clinodactyly of 5th fingers; feet showed brachydactyly and bilateral overlap of the 4th toe over the 3rd and 5th toes, and hallux valgus.
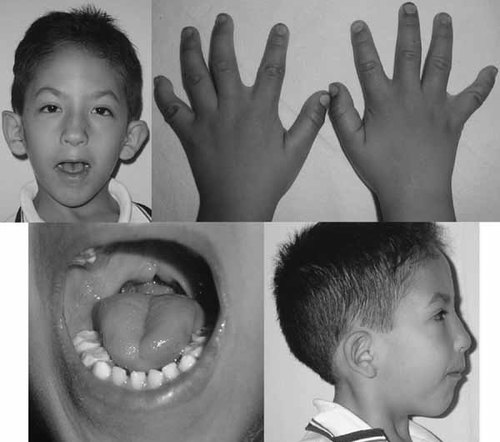
5½-year-old male proband with developmental delay and features of 3p deletion syndrome (also see Table I).
| Features of 3p deletion syndrome | Affected frequencya (%) | Current case |
|---|---|---|
| Growth retardation | 95 | + |
| Ears malformed or low-set | 95 | + |
| Development delay or MR | 81 | + |
| Ptosis | 76 | − |
| Abnormal nose | 76 | + |
| Micrognathia | 68 | + |
| Abnormal philtrum | 68 | + |
| Abnormal muscle tone | 62 | − |
| Hypertelorism | 62 | + |
| Psychomotor retardation | 59 | + |
| Microcephaly | 59 | + |
| Epicanthal folds | 54 | + |
| Thin upper lip | 46 | + |
| Palate abnormalities | 41 | + |
| Polydactyly | 41 | − |
| Sacral dimple | 38 | − |
| Downturned corners of the mouth | 32 | + |
| Upslanting palpebral fissures | 32 | − |
| GI abnormalities | 32 | − |
| Seizures or abnormal EEG | 30 | − |
| Hearing loss | 30 | + |
| Clinodactyly | 30 | + |
| Flat occiput | 24 | + |
| Synophrys or bushy eyebrows | 24 | − |
| Renal abnormalities | 24 | + |
| Cardiovascular abnormalities | 24 | − |
| Triangular face | 19 | + |
| Short neck | 19 | + |
| Retrognathia | 19 | + |
| Brachydactyly | 19 | + |
| Downslanting palpebral fissures | 16 | + |
| Syndactyly | 11 | + |
| Kyphosis or scoliosis | 8 | + |
| Dysplastic teeth | 8 | + |
| Overlap of toes | 5 | + |
| Inverted “V” eyebrows | 3 | + |
| Cytogenetic finding | Various 3p deletions | 45,XY,der(3)t(3;13) (p26.2;q12.13),-13 |
- +, present; −, absent.
- a Frequency data are derived from 37 unique cases reported in the literature [Fineman et al., 1978; Verjaal and De Nef 1978; Gonzalez et al., 1980; Smith and Sachdeva, 1980; Garcia Sagredo et al., 1981; Merrild et al., 1981; Higginbottom et al., 1982; Zergollern and Hitrec, 1983; Beneck et al., 1984; Witt et al., 1985; Reifen et al., 1986; Tolmie et al., 1986; Bueno et al., 1987; Schwyzer et al., 1987; Ramer et al., 1989; Meinecke, 1990; Narahara et al., 1990; Tazelaar et al., 1991; Asai et al., 1992; Nienhaus et al., 1992; Mowrey et al., 1993; Lizcano-Gil and Figuera, 1994; Phipps et al., 1994; Moncla et al., 1995; Warburg et al., 1995; Drumheller et al., 1996; Angeloni et al., 1999; Benini et al., 1999; Wahlström et al., 1999; Kariya et al., 2000; Cargile et al., 2002; Fernandez et al., 2004; Malmgren et al., 2007].
At age 4 years, sensorineural hearing loss and conduction hypoacusis was detected bilaterally by auditory brainstem response (ABR) with greater deficits in the right side; the otoacoustic emissions were diminished by 99% bilaterally. Renal ultrasound revealed a lack of differentiation between medullar and cortical zones as well as diminished size and volume on the left; the right kidney presented with normal morphology. Orthodonic evaluation showed an anterior open bite and superior conic teeth with impaired enamel and caries. Cardiac ultrasound and CT-scan of the brain were normal.
The radiological examination showed delayed osseous development with a fusion between C2 and C3, a mild dorsal scoliosis at T3 through T6 level and a defect in the spinal closure with a bifid spinal type 1 in S1. Table I summarizes the presence or absence of abnormal phenotypic features in the current case compared with their frequencies in 37 unique cases reported in the literature.
MATERIALS AND METHODS
Cytogenetic Analysis
Standard chromosome preparations from all participants were made from PHA-stimulated peripheral blood lymphocytes. Conventional cytogenetic studies of the patient and his parents were carried out on G-banded metaphases. In addition, C-banding of the patient's chromosomes was also performed.
Array-CGH Analysis
Genomic DNA from the patient, mother, and father was isolated from whole blood cells and were used for aCGH analysis by high-resolution, tiled chromosome-specific microarrays (NimbleGen Systems, Madison, WI) to determine copy number variations (CNVs) relative to pooled controls (Sigma–Aldrich, St. Louis, MO). These DNAs were sonicated, randomly labeled with Cy3 and Cy5 9mers, and then hybridized with a MAUI Hybridization System (BioMicro Systems, Salt Lake City, UT) following the Nimblgen aCGH protocols (Nimblegen Systems, Madison, WI). Each chromosome-specific array contains 385,000 isothermal oligonucleotide probes of variable length (50–75mers), adjusted to equalize melting temperaturas (Tm = 76°C). Array oligo sequences were masked for repeats in the genome and the median inter-probe distances were 475 bp (chr 3 array) and 225 bp (chr 13 array).
Hybridized arrays were scanned with a GenePix 4000B Scanner (Molecular Devices Corporation, Sunnyvale, California) and normalized using QSPLINE [Workman et al., 2002] within the NimbleScan software package (Nimblegen Inc., Madison, WI).
The normalized intensities were subsequently analyzed with the Circular Binary Segmentation (CBS) algorithm [Olshen et al., 2004; Venkatraman and Olshen, 2007] to determine the significant breakpoints in log2 intensities along the chromosomes. Using average window sizes of 1x, 5x, 10x, and 20x (x = the median inter-probe distance), we determined the possible segments of the genome that were different between our patients and pooled, population-matched control samples. A segment was considered to be significant if y > 0.3 or y < −0.3 (y = the mean log2 ratio of the probes) over any consecutive number of probes (N) as previously described [Olshen et al., 2004; Venkatraman and Olshen, 2007].
FISH Analysis
FISH analyses of metaphase nuclei were carried out as previously described [Lichter et al., 1990]. Bacterial Artificial Chromosomes (BACs) corresponding to the regions of interest identified on cytogenetic analysis were identified using the UCSC Human Genome Browser at http://genome.ucsc.edu/ (March 2006 freeze) and obtained from an RPCI-11 (Roswell Park Cancer Institute) BAC library. These BAC probes were cultured and the human DNA inserts were extracted. Probes were fluorescently labeled by nick translation and hybridized to metaphase spreads consisting of the patient's lymphocytes. Following hybridization, the nuclei were counterstained with DAPI to distinguish nuclei from background and observed under multi-wavelength fluorescent microscopy.
RESULTS
Cytogenetic Analysis
Using conventional cytogenetic analyses, the karyotype of the patient was initially determined to be 45,XY,der(3)t(3;13)(p25;q11),-13 (Fig. 2). C-banding revealed only one centromere in the abnormal chromosome. The karyotypes of the parents were normal.
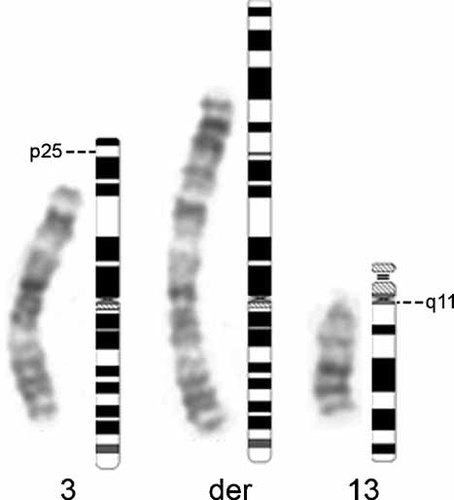
Partial G-banded karyotype of the patient's lymphocytes showing a t(3;13) translocation. Dashed lines indicate the breakpoints on the corresponding ideograms. Shown here is the normal chromosome 3, the derivative chromosome 3 (der), and the normal chromosome 13.
Array-CGH Analysis
Patient DNA was hybridized to two chromosome-specific microarrays, one for chromosome 3 and one for chromosome 13. The log2 ratios of proband DNA hybridization intensities versus pooled controls identified a 3pter deletion with the centromeric extent of this deletion within a 2.7 kb interval spanning base positions 4,510,920–4,513,580, resulting in a terminal deletion of approximately 4.5 Mb (Fig. 3A). The large deletion on chromosome 13, previously identified by cytogenetic analysis, was further defined to have a breakpoint within a 1.3 kb interval on 13q, spanning base positions 26,493,785–26,495,090; this results in a deletion of approximately 26.5 Mb which includes a proximal portion of 13q, the chromosome 13 centromere, and 13p (Fig. 3B). Array results for the parents (not shown) confirm that these deletions in the proband are de novo.
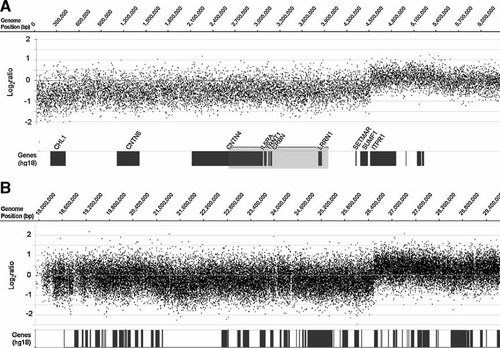
Physical map of the de novo chromosome 3p and chromosome 13q breakpoints in the proband. Array-CGH log2ratios of signal intensity are plotted for each patient probe relative to that of pooled controls along a section of (A) chromosome 3p and (B) chromosome 13q. Base pair (bp) positions are plotted along the X-axis. Locations of known genes based on data at the UCSC Human Genome Browser (http://genome.ucsc.edu/), March 2006 build (hg18), are indicated as dark bars below the CGH data. Abbreviations for genes affected by the deletion of chromosome 3p and the 1.5 Mb critical region for 3p deletion syndrome [Dijkhuizen et al., 2006] (grey shade) are given in (A). The chromosome 3p terminal deletion breakpoint in this proband is approximately 4.5 Mb from the telomere of chromosome 3p. The chromosome 13q breakpoint in this proband is approximately 26.5Mb from the telomere of 13p.
FISH Analysis
To confirm the array CGH results, probes mapping centromeric and telomeric to the presumed translocation breakpoints on chromosome 3p and chromosome 13q in the proband were identified and hybridized to patient lymphocytes (Table II). On the derivative chromosome 3, no fluorescence signal was detected from any BAC probe mapping telomeric to RP11-299N3 (containing the transcript CNTN4) on chromosome 3p or centromeric to RP11-45B20 on chromosome 13, since they fall within the deleted regions (Fig. 4).
| BAC ID | Base positiona | Chromosomal band | Overlapping genes | FISH signal location(s) | Location relative to breakpoint |
|---|---|---|---|---|---|
| RP11-306H5 | chr3:159,320–343,406 | 3p26.3 | CHL1 | nl 3 | telomeric |
| RP11-129K1 | chr3 2,215,034–2,377,366 | 3p26.3 | CNTN4 | nl 3 | telomeric |
| RP11-299N3 | chr3:2,181,279–2,367,269 | 3p26.3 | CNTN4 | nl 3 | telomeric |
| RP11-669E3 | chr3:7,204,187–7,383,435 | 3p26.1 | GRM7 | nl 3, der | centromeric |
| RP11-77P19 | chr13: 17,918,001–18,079,183 | 13q11 | − | nl 13 | centromeric |
| RP11-45B20 | chr13:23,305,109–23,483,639 | 13q12.12 | MIPEP | nl 13 | centromeric |
| RP11-204N9 | chr13:57,108,113–57,207,404 | 13q21.1 | PCDH11 | nl 13, der | telomeric |
- FISH, fluorescence in situ hybridization; BAC, bacteria artificial chromosome; nl 3, normal chromosome 3; nl 13, normal chromosome 13; der, derivative chromosome.
- a Derived from the March 2006 freeze (hg18) of the human genome assembly.

FISH studies of a de novo t(3;13) translocation. A: Two metaphase spreads and two interphase cells. BAC probe RP11-299N3 (red) on chromosome 3p is telomeric to the translocation breakpoint and hybridization signals are seen only on the normal chromosome 3 (nl 3) in both metaphase spreads but not the derivative chromosome 3. One hybridization signal is seen in each interphase cell, confirming hemizygosity in the region of this probe. The probe overlaps with the gene CNTN4 (also see Table II). B: BAC probe RP11-669E3 (red) is centromeric to the translocation breakpoint on chromosome 3p and hybridization signals are seen on the normal chromosome 3 (nl 3) as well as the derivative chromosome 3 (der). This probe overlaps with the gene GRM7. In addition, BAC probe RP11-45B20 (green) on chromosome 13q is within the deleted region, centromeric to the breakpoint, and hybridization signals are seen on the normal chromosome 13q but not the derivative chromosome. Corresponding signals are seen in the inset interphase cell (also see Table II). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
These FISH results confirm the presence of an unbalanced translocation involving a ∼4.5 Mb hemizygous deletion of chromosome 3p as well as a ∼26.5 Mb hemizygous deletion of chromosome 13, which includes a portion of the proximal long arm, the centromere, and entire short arm. Based on these analyses, the translocation pattern in this proband was revealed to be 45,XY,der(3)t(3;13)(p26.2;q12.13),-13.
DISCUSSION
Molecular characterization of rare chromosomal rearrangements and deletions provides a unique opportunity to identify disease genes that may contribute to syndromic phenotypes. Here we present a high-resolution genetic evaluation of a patient with clinical features of 3p deletion syndrome (Tables I and II) using aCGH and FISH. The patient described here has a de novo unbalanced translocation of chromosomes 3 and 13, including monosomy for the terminal portion of 3p, the chromosome 13 centromere, 13p, and a proximal portion of 13q.
While this rearrangement results in haploinsufficiency of multiple genes on both affected chromosomes, it is notable that the deleted genetic material on chromosome 3 overlaps with a recently suggested 1.5 Mb minimal region of overlap for 3p deletion syndrome defined through aCGH analysis of a complex chromosomal aberration in a patient with several features of 3p deletion syndrome [Dijkhuizen et al., 2006] (Fig. 3A). As with other case reports in the literature, the 3p deletion in this patient includes CNTN4, a gene whose disruption has been implicated in causing key aspects of the 3p deletion phenotype, including developmental delay [Fernandez et al., 2004; Dijkhuizen et al., 2006]. Furthermore, variation in CNTN4 has been suggested to contribute toward the etiology of autism spectrum disorders [Fernandez et al., 2004, 2008; Roohi et al., 2008]. While we are unable to make the diagnosis of an autism spectrum disorder based on the available phenotyping of this patient, he did display significant language delay at age 5-1/2, one of three core features in autistic disorder.
With respect to this patient's loss of genetic material on chromosome 13, it is possible that such haploinsufficiency could contribute to certain aspects of this patient's phenotype. However, case reports of partial chromosome 13 deletions suggest that this likely plays a lesser role in his clinical presentation. Deletion of the short arm of the acrocentric chromosome 13 by itself is believed to have little or no consequence and no definitive links between genes in this region and disease phenotypes have been reported [Pires et al., 2005; OMIM, 2008]. A recent comprehensive review of interstitial and terminal 13q deletions suggests that, while varying dysmorphic and mental retardation phenotypes may result from such cases, these are typically the consequence of deletions involving more telomeric aspects of 13q. Centromeric 13q deletions, as seen in the patient reported here, appear to cause fewer anomalies [Ballarati et al., 2007]. However, the literature in this regard is quite sparse, limiting our ability to precisely dissect the relative contributions of chromosome 3 and 13 to the patient's phenotype. Nonetheless, two notable clinical features of our patient are inconsistent with the other case reports of 3p deletion syndrome: a cleft palate/lip and bifid uvula. It is possible that these features may be caused by the deletion on chromosome 13 or by undetected genetic abnormalities elsewhere in the genome.
The patient reported here also has sensorineural hearing loss (SNHL) detected by ABR. SNHL has been described in several cases of 3p deletion syndrome [Higginbottom et al., 1982; Ramer et al., 1989; Narahara et al., 1990; McCullough et al., 2007] but is not a ubiquitous feature. McCullough et al., 2007 compared the extent of 3p deletions in individuals with SNHL and defined a 1.38 Mb minimal candidate region of 3p25.3 in which deletions appear to be associated with this trait. Realizing that cryptic genomic copy number variations (CNVs) may cluster nearby chromosomal breakpoints [Gribble et al., 2005], we evaluated our high-resolution aCGH data to examine this interval for deletions but did not find any putative micro-deletions. Moreover, it is possible that the cause of SNHL in this patient is the deletion on chromosome 13q, as mutations and microdeletions of GHB2 and GJB6 on 13q12 have been associated with nonsyndromic SNHL and these loci are deleted in our patient [del Castillo et al., 2002; Feldmann et al., 2004; del Castillo et al., 2005].
By using FISH and array-CGH, we were able to precisely define a chromosomal rearrangement and deletions in a patient with 3p deletion syndrome. In so doing, we add to the mounting evidence implicating a minimal candidate region including CNTN4, the disruption of which confers key aspects of the 3p deletion syndrome phenotype. This case highlights the utility and unique opportunities provided by the high-resolution investigation of rare chromosomal rearrangements.
Acknowledgements
This project was supported by the Lawrence Family (to M.W.S.). We express our deepest gratitude to the patient and family described herein for their participation in this research. Thank you also to Angeli Landeros-Weisenberger, MD, of the Yale Child Study Center for her assistance in coordinating the initial stages of this collaboration.




