Familial deletion 11q14.3–q22.1 without apparent phenotypic consequences: A haplosufficient 8.5 Mb region†
How to cite this article: Goumy C, Gouas L, Tchirkov A, Roucaute T, Giollant M, Veronèse L, Francannet C, Vago P. 2008. Familial deletion 11q14.3–q22.1 without apparent phenotypic consequences: A haplosufficient 8.5 Mb region. Am J Med Genet Part A 146A:2668–2672.
Abstract
We present the prenatal diagnosis of a chromosome 11q14.3–q22.1 deletion identified in three generations without apparent phenotypic consequences. A 25-year-old G2, P1 woman underwent amniocentesis at 15 weeks' gestation because of a positive result for Down syndrome maternal serum-screening test (1/70). The fetal karyotype revealed an interstitial deletion of the long arm of chromosome 11 confirmed by CGH and FISH: 46,XX,del(11)(q14.3q22.1). The mother and grandfather of the fetus presented the same interstitial deletion with a little if any phenotype effect. The pregnancy was carried to term and resulted in the birth of a normal girl. To our knowledge, only one case of a chromosome 11q14.3–q21 deletion without phenotypic anomalies has been reported. Our study allows the initially described haplosufficient region to be extended from 3.6 Mb to at least 8.5 Mb. This large deletion was compatible with fertility and apparently normal phenotype. Identification of such chromosomal regions is important for prenatal diagnosis and genetic counseling. © 2008 Wiley-Liss, Inc.
INTRODUCTION
Cytogenetically detectable deletions of euchromatic segments are generally expected to result in an abnormal phenotype. There is, however, a small group of several chromosomal regions that upon deletion do not seem to cause an abnormal phenotype [Barber, 2005; Kowalczyk et al., 2007]. Identification of such deletions is essential for prenatal diagnosis and genetic counseling. Here we report on the second-trimester prenatal diagnosis of an interstitial 11q14.3–q22.1 deletion with direct transmission in three generations without apparent phenotypic effect. The complete characterization of the abnormality by CGH and FISH as well as the control analysis of parental chromosomes were performed. Finally, we consider the possible contributory factors which might explain the lack of clinical significance of this large deletion.
CLINICAL REPORT AND CYTOGENETIC STUDY
The patient was a G2P1 25-year-old woman, who had amniocentesis because of a positive Down syndrome maternal serum-screening result at 15 weeks' gestation. Ultrasound scans at 11 and 15 weeks gestation were normal. The family history was unremarkable.
Chromosome analysis was performed on amniocyte cultures with GTG-banding according to standard methods and revealed a deletion of the long arm of chromosome 11 (Fig. 1a).
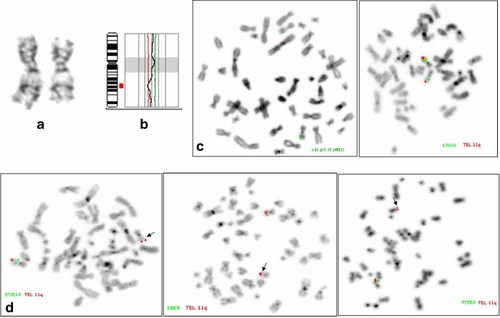
a: Partial GTG-banded karyotype of the fetus illustrating normal (left) and deleted (right) chromosome 11 homologues. b: CGH profile of chromosome 11 showing the 11q14.3 → 11q22.1 deletion. c: FISH with the Vysis API2 probe localized at 11q22.22 and a BAC clone RP11-12D16 localized at 11q14.2 showed two green signals. d: FISH with BAC clones mapping to 11q14.3 (RP11-372E19), 11q21 (RP11-16K5), and 11q22.1 (RP11-775E2) showed only a single copy of these probes (arrows). The subtelomeric 11q probe (Vysis) was used as a control (red signals). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Molecular Cytogenetic Analysis
Comparative genomic hybridization (CGH) was performed on chromosome spreads using DNA extracted from cultured amniocytes, as previously described [du Manoir et al., 1995; Goumy et al., 2005], with a resolution of 3–5 Mb determined in our laboratory. The CGH confirmed the 11q deletion and mapped the breakpoints at 11q14.3 and 11q22.1 (Fig. 1b). FISH (fluorescence in situ hybridization) experiments were performed on metaphase spreads with a commercial probe (LSI API2, Vysis) according to the manufacturer's instructions, and with bacterial artificial chromosome (BAC) selected from the human library RPCI-11 according to the UCSC Human Genome Assembly (March 2006). BAC DNA extraction was done using the NucleoSpin® Plasmid kit (Macherey-Nagel GmbH, Hoerdt, France). BAC DNAs were then labeled with Fluorescein-12-dUTP (Roche Diagnostics GmbH, Meylan, France) by nick translation (Abbott Molecular, Rungis, France).
FISH results with a BAC clone localized at 11q14.2 (RP11-12D16) showed two signals, confirming a breakpoint location between 11q14.2 and 11q14.3, whereas the commercial probe LSI API2 at 11q21 also showed two signals which was not concordant with the 11q22.1 breakpoint shown by CGH (Fig. 1c). FISH with BAC clones localized at 11q14.3 (RP11-372E19), 11q21 (RP11-16K5), 11q22.1 (RP11-775E2) showed one signal on one of the chromosome 11 homologues (Fig. 1d) and made it possible to confirm that the deletion extended to the 11q22.1 band. Using the University of California Santa Cruz (UCSC) genome browser (http://genome.ucsc.edu) with the last genome assembly (NCBI Build 36.1, March 2006), we were able to replace the position of the commercial probe LSI API2 from 11q21, as given by the manufacturer, to 11q22.2. This new localization explained the presence of two FISH signals with this probe and confirmed the breakpoint in 11q22.1.
By combining karyotype, CGH and FISH results, the karyotype was described as follows: 46,XX.ish del(11)(q14.3q22.1)(RP11-372E19-,RP11-16K5-,RP11-775E2-). The minimal deletion size estimated as the distance between the proximal edge of BAC RP11-372E19 and the distal edge of BAC RP11-775E2 was 8.5 Mb and the maximal deleted size evaluated between the distal edge of RP11-12D16 and the proximal edge of the LSI API2 probe was 16 Mb (Fig. 2).
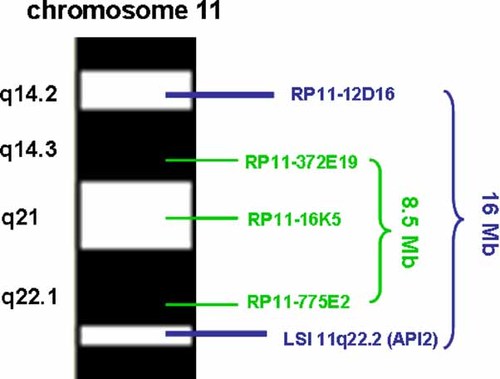
Partial chromosome 11 ideogram showing the BAC clones used to define the deletion. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Familial Study
Chromosome analyses were performed on both parents who apparently were phenotypically normal. The same interstitial deletion was observed in the mothers' karyotype. CGH analysis showed the same profile with breakpoints at 11q14.3 and 11q22.1 and similar FISH results were obtained with the BAC clones used in the fetus (data not shown). The mother's clinical examination and interrogation revealed endometrial polyps (which were treated by hysteroscopic surgery), toe camptodactyly and ophtalmologic disorders (hypermetropia, astigmatism, strabismus). There was no evidence of multiple miscarriages, birth defects or developmental delay in the mother's family.
The maternal grandparents were then explored, and cytogenetic analysis in the phenotypically normal grandfather revealed the 11q14.3q22.1 deletion (data not shown).
Pregnancy Issue
The genetic counseling was reassuring. The pregnancy was carried to term and resulted in the birth of a normal girl without dysmorphic features. Birth weight was 3,400 g, length 50 cm and head circumference 35 cm. The Apgar scores were 8/8/10 and clinical examination was normal.
DISCUSSION
This case provides further evidence that rare unbalanced deletions of specific chromosomal regions may result in the absence of phenotypic effect and underlines the importance of obtaining parental karyotypes before making a clinical decision. The interstitial deletion 11q14.3–q22.1 described here was identified in three individuals from three generations without apparent phenotypic consequences. The camptodactyly of the toes and ophthalmological features observed in the mother were not present in the grandfather. These features are common in the general population and are unlikely to be related to the deletion.
In 2007, Kowalczyk et al. reviewed cases of euchromatic chromosome imbalances, mainly inherited, without phenotypic abnormalities. They found 15 chromosomal regions on chromosomes 2, 3, 5, 7, 8, 9, 10, 11, 13, 16, and 18 for which deletions are observed in phenotypically normal individuals. Some of these, namely deletions 5p14 [Overhauser et al., 1986; Johnson et al., 2000], 10q11 [Bisgaard et al., 2007], 16q21 [Hand et al., 2000], and 2p12 [Barber, 2005] have each been found in more than one independent family and transmitted without consistent phenotypic consequences. In the review by Barber 2005, the average size of such deletions was 8.2 Mb (range 4.2–16 Mb) and these deletions generally involved G dark bands to which few genes map.
To our knowledge, only one asymptomatic deletion of the long arm of chromosome 11 has been described [Li et al., 2002]. This report identified a 3.6 Mb deletion at 11q14.3–q21 in five individuals representing three generations of kindred. Only one carrier showed mild clinical features, which were probably unrelated to the deletion.
In our case, the deletion was visible on standard karyotype with a resolution of about 10 Mb. FISH with commercial and BAC probes mapped the breakpoints at 11q14.3 and 11q22.1 and made it possible to estimate the size of the deletion between 8.5 and 16 Mb. Thus, our report enables the haplosufficient region initially described by Li et al. to be extended from 3.6 Mb to at least 8.5 Mb. This 8.5 Mb region contains only 22 protein coding genes (Table I; http://www.ncbi.nlm.nih.gov/mapview).
| Gene symbol | Gene name | Expression | Gene/protein family | Function | Related disease | Other gene family location |
|---|---|---|---|---|---|---|
| FAT3* | Tumor suppressor homolog 3 (drosophila) | ES cells, primitive neuroectoderm, fetal brain, infant brain, adult neural tissues and prostate | Cadherin superfamily members | Cell adhesion | None | FAT1 at 4q35, FAT2 at 5q32q33, FAT4 at 4q28.1 |
| MTNR1Ba/MEL-1B-R | Melatonin receptor 1B | Retina, brain | Receptor for melatonin | Receptor for melatonin | 600804 | MTNR1A at 4q35.1 |
| SLC36A4/PAT4 | Solute carrier family 36 | Unknown | Amino acid transporter genes | Proton-coupled transporters | None | Cluster at 5q33.1 |
| JOSD3/MGC5306 | Josephin domain containing 3 | Only in human carcinoma and tumor cell lines | Unknown | Repair pathway in carcinogenesis | None | 22q13.1 19q13.33 |
| MED17/CRSP6/TRAP80 | Mediator complex subunit 17/Cofactor required for Sp1 | Ubiquitously | Transcription cofactor complex CRSP | Cofactor required for transcription initiation by the RNA pol II | 603810 | Multiple |
| HEPHL1 | Hephaestin-like | Unknown | Unknown | Bind copper Feroxydase activity Intestinal iron absorption | None | Unknown |
| PANX1/MRS1 | Pannexin 1 | Ubiquitously abundantly in several brain regions and retina | Innexin/pannexin superfamily | Structural components of gap junctions | 608420 | PANX2: 22q13.33 PANX3: 11q24.2 |
| GPR83/GIR | G protein coupled receptor 83 | Brain | G protein-coupled receptors | Unknown | 605569 | 2p14 |
| MRE11A | Meiotic recombination 11 homolog A | Ubiquitously High level in proliferating tissues | MRE11 family | Homologous recombination, telomere length maintenance, DNA DSBs repair | 600814 | MRE11B at 3q25 |
| ANKRD49/FGIF | Ankyrin repeat domain 49 | Unknown | Unknown | Unknown | None | Unknown |
| FUT4 | Fucosyltransferase 4 | Embryos (5–10 weeks) | Fucosyltransferases | Biosynthesis of Lewis antigene | 104230 | Multiple FUT9: 6q16 |
| PIWIL4 | Piwi-like 4 (drosophile) | Adult testis | PIWI-like family | Development and maintenance of germline stem cells | 610315 | 12q24.33, 8p21.3, 22q11.23 |
| AMOTL1 | Angiomotin like 1 | Endothelial cells of capillaries and placenta vessels | AMOTL2 | Structural component of tight junctions | None | 3q21q22 |
| JMJD2D | Jumonji domain containing 2D | Prostate | JMJD2 family | Histone demethylase/Androgen receptor activators | 609766 | 1p34.1, 9p24.1, 19p13.3 |
| SFRS2B/SRP46 | Splicing factor, arginine/serine-rich 2B | Multiple tissues | Serine/arginine-rich family | Pre-mRNA Splicing factor | 603269 | Multiple |
| ENDOD1 | Endonuclease domain containing 1 | Unknown | Unknown | Unknown | None | Unknown |
| SESN3 | Sestrin 3 | Unknown | Sestrin family/PA26-related gene family | Unknown | 607768 | 1p35.3 6q21 |
| CEP57/PIG8 | Centrosomal protein 57kDa/translokin | Ubiquitinously | Unknown | Mediates FGF2 nuclear translocation and mitogenic activity | 607951 | Unknown |
| MTMR2/CMT4B | Myotubularin related protein 2 | Ubiquitinously | Myotubularin related family (14 members) | Tyrosine phosphatase | 603557 | Xq28, 13q12 |
| MAML2/MAM2 | Mastermind-like2 | Mastermind proteins family | NOTCH coactivator signaling pathway | 607537 | 5q35, 4q28 | |
| JRKL/HHMJG | Jerky homolog-like (mouse) | Ubiquitinously | Unknown | Nuclear regulatory protein | 603211 | Unknown |
| CNTN5/NB-2 | Contactin 5 | Developing nervous system, brain moderate in thyroid and placenta | Immunoglobulin superfamily | Cell adhesion molecule | 607219 | 12q11q12, 1q32.1, cluster at 3p26 |
- * From Human Genome Assembly Build 36 (March 2006).
- a FAT1-MTNR1A locus at 4q35.2 and FAT3-MTNR1B locus at 11q14.3–q21 were paralogous regions (Katoh and Katoh, 2006).
Stratton et al. 1994 described a chromosome11q14.1–q21 deletion in a 4-year-old girl with moderate developmental delay, horseshoe kidney, bilateral duplication of the ureters with right upper pole obstruction, hydronephrosis and Wilms tumor. In this case, the 11q14.3–q21 region of deletion overlaps with the haplosufficient region described here, suggesting that the genes responsible for the abnormal phenotype may be localized between 11q14.1 and 11q14.3.
Horelli-Kuitunen et al. 1999 described a de novo deletion of 11q21–q22.3 in a 3-year-old girl with disproportionate short stature, hypotonia with “myopatic changes” at the muscle biopsy, developmental delay, hypertelorism, low-set ears, pes equinovarus of the right foot, mild hydronephrosis, enlarged cerebral lateral ventricles and thin corpus callosum. Meyer et al. 2000 reported a 11q21–q23.1 deletion in a 21-year-old woman associated with motor and speech developmental delay in infancy, congenital heart defect, craniofacial anomalies, menstrual irregularity, hirsutism, elevated serum androgen levels and polycystic ovary syndrome. Likewise, Li et al. 2006 reported an 11q14.1–q23.2 deletion and predicted that the distal segment 11q21–q23.3 may be responsible for the varying degrees of growth retardation, mental retardation, cleft palate and minor digital anomalies of the patient. Assuming that the 11q14.3–q22.1 region might be haplosufficient, the regions responsible for the abnormal phenotype in these three cases could be reduced to 11q22.1–q22.3, 11q22.1–q23.1, and 11q22.1–q23.2, respectively.
The hypotheses proposed to explain the lack of clinical consequences in case of such large deletions were the low density of genes in the deleted region, the haplosufficiency of the majority of these genes or the presence of genes with similar function located elsewhere in the genome and a possible genomic imprinting effect leading to the loss of inactive material [Li et al., 2002; Barber, 2005].
Our observation does not support the genomic imprinting hypothesis [Bortotto et al., 1990; Li et al., 2002] because the transmission of the deletion in three generations of apparently normal individuals was of both paternal and maternal origin, even if the psychomotor development of the newborn is difficult to assess.
The majority of the deleted genes have related loci on other chromosomes, suggesting that functional complementation can arise from isoform expression or that alternative metabolic pathways can substitute those affected (Table I). The other genes are probably not dosage sensitive.
Recently, identification of inherited and de novo imbalances of uncertain clinical significance has progressively escalated with the increasing use of array technology in diagnostic laboratories. Knowledge of such harmless imbalances is particularly helpful for prenatal diagnosis and genetic counseling.




