Novel SCN1A frameshift mutation with absence of truncated NaV1.1 protein in severe myoclonic epilepsy of infancy†
How to cite this article: McArdle EJ, Kunic JD, George AL Jr. 2008. Novel SCN1A frameshift mutation with absence of truncated NaV1.1 protein in severe myoclonic epilepsy of infancy. Am J Med Genet Part A 146A:2421–2423.
To the Editor:
Severe myoclonic epilepsy of infancy (SMEI) or Dravet syndrome (OMIM 607208) is a severe epileptic encephalopathy with onset during the first year of life with devastating neurological sequelae [Dravet et al., 1992; Wolff et al., 2006]. Typically, SMEI presents during infancy with febrile seizures often followed by a prolonged period of drug-refractory epilepsy with repeated bouts of status epilepticus. Patients exhibit early developmental delays, progressive cognitive impairment, and often ataxia. Most reported cases are sporadic but a family history of seizures may be present [Kimura et al., 2005].
SMEI has been associated with a large number (>150) of predominantly de novo mutations in SCN1A encoding the brain voltage-gated sodium channel NaV1.1 [Meisler and Kearney, 2005; Mulley et al., 2005]. Approximately half of reported SCN1A alleles associated with SMEI cause protein truncation either by non-sense or frameshift mutation. Further, a large proportion of missense mutations studied in vitro confer a loss-of-function phenotype to the channel protein [Sugawara et al., 2003; Rhodes et al., 2004; Ohmori et al., 2006]. These findings suggest that haploinsufficiency is a plausible mechanism to explain the disease and this idea is consistent with the observations that mice heterozygous for either a null or truncated Scn1a allele exhibit a severe seizure phenotype [Yu et al., 2006; Ogiwara et al., 2007].
Alternatively, it is conceivable that truncating mutations in SCN1A may cause seizures through dominant-negative effects imposed by the mutant protein on the wild-type (WT) allele, if the mutant protein is expressed. Kamiya et al. 2004 identified a non-sense SCN2A mutation (p.R102X) in a patient with a clinical syndrome resembling SMEI. These investigators used in vitro heterologous expression studies to demonstrate that the truncated protein exerts a dominant-negative effect on the WT channel. Unfortunately, this study could not assess whether the predicted truncated protein was expressed in vivo as this is a common challenge due to the usual absence of brain tissue from SMEI subjects.
Here we report on a unique opportunity to study post-mortem brain tissue obtained from a child affected by SMEI, and a direct assessment of whether the pathogenesis is more readily attributable to haploinsufficiency or a dominant-negative effect. The patient had her first seizure at age 6 months. She exhibited subsequent developmental delays and was diagnosed with SMEI at age 2 years; she died due to complications of the disorder at the age of 5 years.
We obtained frozen post-mortem cerebellar tissue from the patient and from an age/gender/race-matched control subject. A second control post-mortem cerebellar sample from a 24-year-old male subject. All samples were obtained from tissue banks and were anonymous; no additional information regarding medical history or DNA samples of first degree relatives were available. Total RNA was isolated from frozen tissue, followed by first strand cDNA synthesis. The patient's complete SCN1A coding sequence (GenBank AB093548) was amplified in four overlapping segments as described previously [Lossin et al., 2002], and sequence analysis revealed a heterozygous variant, c.3608delA, which predicts p.Gln1203HisfsX4 (Fig. 1a). Both the nucleotide deletion and the predicted pre-mature stop codon occur within the intracellular cytoplasmic linker region between domains D2 and D3 of NaV1.1 (Fig. 1b).
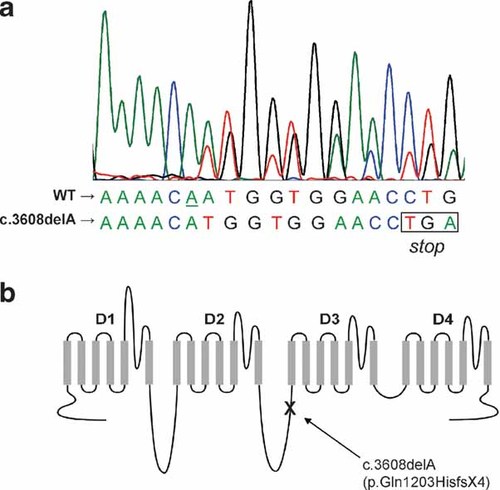
Identification of a novel SCN1A mutation in SMEI. a: Nucleotide sequence electropherograph of cDNA isolated from the SMEI subject indicating a single-allele deletion of an adenosine (underlined) at nucleotide position 3,608 resulting in a frameshift with a pre-mature stop codon (boxed) introduced at amino acid position 1,207. b: Location of the mutation (c.3608delA) and predicted frameshift in the sodium channel topology.
We next assessed the expression of NaV1.1 protein in the cerebellum of the SMEI patient and the two controls by Western blotting, using an anti-NaV1.1 antibody raised against a NaV1.1-specific epitope (residues 465–481, Alomone Labs, Jerusalem, Israel). Importantly, this epitope is preserved in the predicted truncated NaV1.1 protein and therefore the primary antibody should be capable of immunoreacting with both the full-length and mutant proteins. With normalized total protein input, Western blot analysis for NaV1.1 performed on protein lysates prepared from the three cerebellar samples reproducibly demonstrated only the full-length protein (260 kDa) in lysates from the SMEI case and both controls, but the predicted truncated protein (predicted molecular mass ∼136 kDa) was never observed (Fig. 2a) even after prolonged exposures.
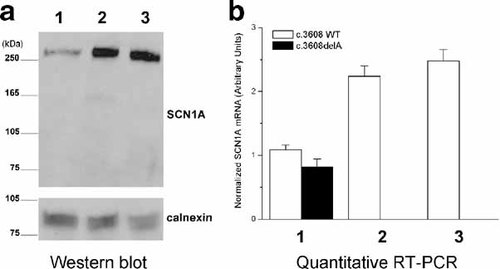
Truncated SCN1A protein isoform not detected in cerebellum. a: Immunoblot for NaV1.1 (upper panel labeled as SCN1A) in protein lysates from cerebellum of SMEI subject (lane 1), age/gender/race- matched control (lane 2), and adult male (lane 3). Immunodetection of calnexin (lower panel) was performed to assure equal protein content of the samples. b: Quantitative real-time RT-PCR for wild-type (WT) and mutant (c.3608delA) mRNA transcripts in SMEI and control brain samples (same lane order as in a). In lane 1, differences between WT and c.3608delA are significant (P < 0.05). Differences in WT transcript levels between lane 1 and both controls were significant at the P < 0.0001 level. Statistical comparisons were made using one-way ANOVA followed by a Tukey post-test.
Absence of the predicted truncated protein might be explained by non-sense mediated decay of the mutant mRNA transcript. To investigate this possibility, we measured levels of the wild-type (WT) and mutant transcripts with a quantitative real-time RT-PCR allele discrimination assay. We used PrimerExpress 2.0 software (Applied Biosystems, Foster City, CA) to design amplification primers that do not hybridize with genomic DNA (3516F 5′-AGAAGCTTGTTTCACTGAAGGC-3′, 3697R 5′-TCAAATGCCAGAGCACCAC-3′) and probes that discriminate the WT and mutant alleles based on the single nucleotide deletion at c.3608 (c.3608-WT VIC-TTCCACCATTGTTT-MGB, c.3608delA FAM-TTCCACCATGTTT-MGB; MGB, minor grove binding molecule). For each tissue, results were compared to an allele-specific standard curve and normalized to expression of GAPDH, assayed in the same samples. Cycling was performed on an Applied Biosystems 7900HT; each sample was run in triplicate on two separate occasions for a total of six replicates.
Figure 2b illustrates that both alleles were detected in cDNA from SMEI cerebellum but only WT was observed in the two controls. Importantly, the level of WT mRNA in the SMEI tissue was approximately half of the levels observed in the controls but was slightly greater than the c.3608delA transcript (P < 0.05). These results indicate that the frameshift mutation does not greatly impair the level of steady-state c.3608delA mRNA and suggest that the lack of truncated protein encoded by this mutant allele cannot be explained by absence of the corresponding mRNA transcript.
Truncated proteins are likely to be mis-folded or mis-assembled in the endoplasmic reticulum (ER) and subject to ER quality-control mechanisms that may target aberrant proteins for ER-associated degradation (ERAD) [Sanders and Myers, 2004; Meusser et al., 2005]. We speculate that ERAD explains the absence of the predicted truncated NaV1.1 protein in this case. We acknowledge an alternative possibility, that the predicted truncated protein may be more susceptible to post-mortem degradation than full-length NaV1.1, and in an extreme case the predicted truncated protein could exist in vivo yet elude detection by the methods employed here. However, we believe this scenario is less likely than that of a known cellular mechanism (ERAD) accounting for the absence of the predicted truncated protein.
In summary, we identified a novel SCN1A frameshift mutation associated with SMEI and investigated both mRNA and protein expression in brain tissue from the affected mutation carrier. Our findings indicate that expression of a predicted truncated protein resulting from a SCN1A frameshift mutation should not be assumed and that haploinsufficiency remains the most likely disease mechanism.
Acknowledgements
We acknowledge the NICHD Brain and Tissue Bank for Developmental Disorders at University of Maryland and the Harvard Brain Tissue Resource Center for providing human tissues.




