A previously unrecognized microdeletion syndrome on chromosome 22 band q11.2 encompassing the BCR gene†
How to cite this article: Mikhail FM, Descartes M, Piotrowski A, Andersson R, Diaz de Ståhl T, Komorowski J, Bruder CEG, Dumanski JP, Carroll AJ. 2007. A previously unrecognized microdeletion syndrome on chromosome 22 band q11.2 encompassing the BCR gene. Am J Med Genet Part A 143A:2178–2184.
Abstract
Susceptibility of the chromosome 22q11.2 region to rearrangements has been recognized on the basis of common clinical disorders such as the DiGeorge/velocardiofacial syndrome (DG/VCFs). Recent evidence has implicated low-copy repeats (LCRs); also known as segmental duplications; on 22q as mediators of nonallelic homologous recombination (NAHR) that result in rearrangements of 22q11.2. It has been shown that both deletion and duplication events can occur as a result of NAHR caused by unequal crossover of LCRs. Here we report on the clinical, cytogenetic and array CGH studies of a 15-year-old Hispanic boy with history of learning and behavior problems. We suggest that he represents a previously unrecognized microdeletion syndrome on chromosome 22 band q11.2 just telomeric to the DG/VCFs typically deleted region and encompassing the BCR gene. Using a 32K BAC array CGH chip we were able to refine and precisely narrow the breakpoints of this microdeletion, which was estimated to be 1.55–1.92 Mb in size and to span approximately 20 genes. This microdeletion region is flanked by LCR clusters containing several modules with a very high degree of sequence homology (>95%), and therefore could play a causal role in its origin. © 2007 Wiley-Liss, Inc.
INTRODUCTION
Contiguous gene syndromes represent one group of genomic disorders that result from either deletions or duplications of chromosomal segments that encompass several adjacent dosage-sensitive genes. The result is a copy-number variation (either loss or gain) of these genes [Schmickel, 1986]. The vast majority of these syndromes result from submicroscopic chromosomal rearrangements, therefore limiting their detection by conventional cytogenetic analysis; including high-resolution chromosome analysis; hence they have been referred to as microdeletions and microduplications. However, the application of fluorescence in situ hybridization (FISH) analysis to either metaphase or interphase chromosomes has allowed more sensitive detection of these subtle rearrangements. The genomic segments involved in several of these conditions have been shown to be flanked by low copy repeats (LCRs); also known as segmental duplications; that share a very high degree of sequence homology (>95%) and the rearrangements to be mediated by non-allelic homologous recombination (NAHR) [Stankiewicz and Lupski, 2002].
Chromosome 22 band q11.2 has been recognized to be highly susceptible to these subtle microdeletions and microduplications. These rearrangements were initially recognized on the basis of common clinical disorders such as the DiGeorge/velocardiofacial syndrome (DG/VCFs). Recent evidence has implicated LCRs on 22q11.2 as mediators of NAHR that result in these rearrangements. Both deletion and duplication events of the DG/VCFs critical region have been described [Edelmann et al., 1999; Shaikh et al., 2000; Ensenauer et al., 2003]. At least eight LCR clusters have been identified on chromosome band 22q11.2 and have been named LCR22s [Edelmann et al., 1999]. Only four of these have been implicated in the origin of the DG/VCFs typically deleted region, and have been termed LCR22-A, LCR22-B, LCR22-C and LCR22-D (ordered from centromere to telomere) [Edelmann et al., 1999; Shaikh et al., 2000]. The 22q11.2 microdeletion size range from ∼3.0 Mb (>90% of DG/VCFs patients) to ∼1.5 Mb, with no evidence of involvement of LCR clusters telomeric to LCR22-D [Carlson et al., 1997; Edelmann et al., 1999]. Conversely, 22q11.2 microduplications as large as ∼6.0 Mb that span the DG/VCFs critical region have been reported and involve LCR clusters telomeric to LCR22-D. It has been concluded that deletions more than 3.0 Mb in this genomic region are probably not viable [Ensenauer et al., 2003]. Recently Ravnan et al. in a subtelomere FISH study of 11,688 cases with developmental disabilities reported eight cases with an interstitial deletion on 22q11.2 in the region just telomeric to the DG/VCFs critical region and encompassing the BCR gene. The clinical phenotypes of these eight cases were discussed only briefly [Ravnan et al., 2006].
Here we report on the clinical, cytogenetic and array CGH studies of a 15-year-old Hispanic boy with history of learning and behavior problems. We suggest that he represents a previously unrecognized microdeletion syndrome on chromosome 22 band q11.2 just telomeric to the DG/VCFs typically deleted region and similar to the eight cases reported by Ravnan et al. 2006.
PATIENT AND METHODS
Clinical Report
Our patient was born at 35 weeks gestation by Caesarian section in Mexico. He is the first child born to a 25-year-old G1 mother. His birth weight was 2.45 kg. There was no history of maternal illness. The patient had no history of any birth defects and parents denied any complications during the newborn period. His developmental milestones were within normal limits. The patient was noted to have problems when he started school and was diagnosed in Mexico with attention deficit-hyperactivity disorder (ADHD). His parents moved to the USA when he was 8 years old. A recent evaluation showed that the patient's receptive language skills are normal for his age, but his performance on the visual-motor integration (VMI) test yielded an age equivalent of 8.4 years.
The patient was first examined in our clinic at age 15 years. On physical examination he was shy but very cooperative. He weighed 83.1 kg (98th centile), height was 159.7 cm (10th centile), and occipitofrontal circumference was 58.6 cm (>95th centile). He displayed slight facial dysmorphism, including short and narrow forehead, flared eyebrows, upslanting palpebral fissures, short philtrum, long pointed nose, intact palate, cupped ears with small lobules and short neck (Fig. 1). Thorax was symmetrical and clear on auscultation. Heart was normal, and abdomen was soft, depressible with no apparent organomegaly. The back was intact. He had a normal muscle mass and normal skin. His extremities showed mild tapering of the fingers with bilateral 5th finger clinodactyly.

The patient at age 15 years. Note the short and narrow forehead, flared eyebrows, up-slanting palpebral fissures, short philtrum, long pointed nose, cupped ears with small lobules and short neck.
Cytogenetic and Fluorescence In Situ Hybridization (FISH) Analyses
Routine as well as high-resolution GTG-banding chromosome analyses and FISH analysis were performed on metaphase preparations of peripheral blood lymphocytes from the patient and his parents using standard techniques. The chromosomes were analyzed and the karyotype described according to the International System for Cytogenetic Nomenclature (ISCN 2005) [Shaffer and Tommerup, 2005].
The FISH probes used included the TelVysion panel of subtelomeric probes (Vysis, Downers Grove, IL), the LSI BCR probe (22q11.2) contained in the BCR-ABL single fusion translocation probe set (Vysis), and the Tuple1 probe (22q11.2) within the DG/VCFs critical region (Vysis). The LSI BCR probe (22q11.2) is approximately 300 kb, spans the centromeric three fourths and extends beyond the 5′ end of the gene (Fig. 4). We also used the LSI BCR dual fusion probe (22q11.2) contained in the BCR-ABL dual fusion translocation probe set (Vysis), which is composed of two probes each approximately 600 kb with a 300 kb gap in between. The centromeric portion spans the BCR gene (Fig. 4). Slide hybridization and washes were performed using standard FISH protocols. The slides were then counter stained with DAPI and analyzed with an Olympus BX61 microscope (Olympus America, Inc., Melville, NY) equipped with the appropriate filter combination and a CCD camera, and coupled to the CytoVision image analysis system (Applied Imaging, Santa Clara, CA). Fifteen metaphases were scored for the BCR and Tuple1 probe sets, whereas four metaphases were scored for each of the subtelomere probe sets.
32K BAC Array CGH Study
Design of DOP-PCR 32K BAC Array
The array was constructed using the human BAC minimal tiling clone set which were purchased from CHORI (http://bacpac.chori.org/pHumanMinSet.htm). This clone set provides complete coverage of sequenced parts of the human genome with average resolution of 100 kb. DNA isolated from the clones was DOP-PCR amplified [Fiegler et al., 2003] and spotted on HD CodeLink slides (GE Healthcare Bio-Sciences, Piscataway, NJ) with a Microarray Printer constructed by the bioinstrumentation group at the Lawrence Berkeley National Laboratory, Berkeley, CA. Prior to analysis, the array was validated extensively using previously well characterized samples including: (i) 71 phenotypically normal controls (44 men and 27 women) from three different ethnic populations (Asians, Africans and Caucasians); (ii) NF2 patient carrying a 22q deletion; (iii) NF1 patient carrying the typical 1.5 Mb deletion on 17q; (iv) Charcot-Marie-Tooth patient carrying a 17p duplication; (v) Tumor tissue derived DNA from glioblastoma multiforme and meningioma samples; (vi) Human small cell lung cancer cell line U2020 and human nasopharyngeal carcinoma cell line HONE1 (Bruder et al., personal communication; Piotrowski et al., personal communication; Diaz de Ståhl et al., personal communication).
Hybridization, scanning and data analysis
The methods used for blocking CodeLink slides, DNA labeling, and post-hybridization processing have been described in detail elsewhere (Mantripragada et al., 2006). Briefly, the slides were blocked using sodium boro-hydride treatment, and pre-hybridized (5× SSC, 0.1% SDS, 0.4% BSA; for 1 hr at 45°C). Test and reference DNA were labeled by random priming with Cy3-dCTP and Cy5-dCTP (GE Healthcare Bio-Sciences) respectively, using Bioprime Array CGH Genomic Labeling System with purification module (Invitrogen, Carlsbad, CA). Labeled DNA was suspended in the hybridization buffer (2× SSC, 4% SDS, 50% formamide, 10% dextran sulfate) and hybridized to the array for 20 hr at 45°C. Post-hybridization processing consisted of four washing steps: (i) 2× SSC, 0.1% SDS, 25% formamide for 20 min at 45°C; (ii) 1× PBS for 10 min at room temperature (RT); (iii) 0.2× SSC for 15 sec at RT; (iv) Deionized water for 5 sec at RT. The slides were dried with compressed air. Image acquisition was performed using the Axon 4000B scanner (Molecular Devices, Sunnyvale, CA). Analysis of hybridization intensity was carried out using the GenePix Pro 6.0 image analysis software (Molecular Devices). Subsequent data analysis steps were performed in the Linnaeus Center for Bioinformatics, Data WareHouse, Uppsala University, Sweden (http://dw.lcb.uu.se). The steps of data processing were as follows: filtering out the spots containing >5% of oversaturated pixels, normalization of data by print-tip LOESS method, discarding the spots which showed signal-to-noise ratio less than three in both Cy3 and Cy5 channels, filtering out the measurement points which were marked “bad” or “empty” in GenePix Pro. The normalized median fluorescence intensity Cy3/Cy5 ratios were used for graphical representation of data.
RESULTS
Cytogenetic analysis of 15 GTG-banded metaphase spreads in the patient showed a normal male karyotype. Subtelomeric FISH analysis showed normal subtelomeric regions, but TelVysion subtelomere mixture no. 3 containing the 22q subtelomere probe showed an interstitial deletion of the LSI BCR control probe (22q11.2) (Fig. 2B). The LSI BCR control probe in the subtelomere mixture no. 3 is the exact same probe contained in the BCR-ABL single fusion translocation probe set. These results were confirmed using the BCR-ABL single fusion translocation probe set. Using the BCR-ABL dual fusion translocation probe set only showed a reduced signal size on the deleted chromosome 22 (Fig. 2C). In order to check the integrity of the DG/VCFs critical region, we used the Tuple1 probe (22q11.2), which showed a normal hybridization pattern indicating that the DG/VCFs critical region was intact (Fig. 2D). A map of all the FISH probes used is shown in Figure 4. Both parents proved to be normal cytogenetically both by GTG-banded high-resolution chromosome and FISH analyses. The patients final karyotype was: 46,XY.ish del(22)(q11.2q11.2)(BCR-)dn.
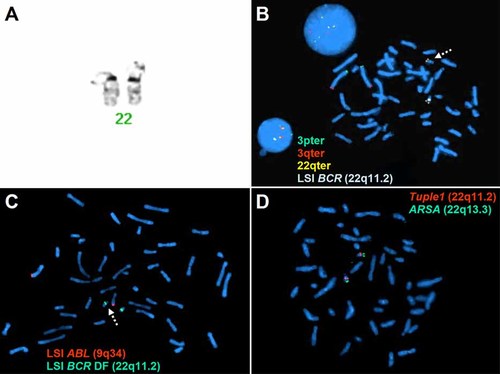
Cytogenetic and FISH analyses of the patient. A: Partial karyotype showing GTG-banded chromosome 22. B: Metaphase and interphase FISH using TelVysion mixture no. 3 set of subtelomeric probes, which includes 3pter (labeled in SpectrumGreen), 3qter (labeled in SpectrumOrange), 22qter (labeled in SpectrumGreen and Orange), as well as LSI BCR control probe (22q11.2) (labeled in SpectrumAqua). Note the deletion of the BCR probe signal (dashed arrow). C: Metaphase FISH using the LSI BCR-ABL dual fusion translocation probe, which includes LSI BCR dual fusion probe (22q11.2) (labeled in SpectrumGreen) and LSI ABL probe (9q34) (labeled in SpectrumOrange). Note the reduced BCR probe signal size (dashed arrow). D: Metaphase FISH using Tuple1 probe (22q11.2) (labeled in SpectrumOrange) and mixed with ARSA control probe (22q13.3) (labeled in SpectrumGreen). Note the intact Tuple1 probe signal on both chromosomes 22. [Color figure can be viewed in the online issue, which is available at http://www.interscience.wiley.com.]
Using the 32K BAC array CGH chip we were able to precisely map both the proximal and the distal breakpoints of the deletion. The proximal breakpoint of the deletion mapped to 20.15–20.37 Mb between clones CTD-2295P14 and RP11-794K20, whereas the distal breakpoint mapped to 21.92–22.07 Mb between clones RP11-61N10 and RP11-529P21 (Fig. 3). Therefore this interstitial microdeletion was estimated to be 1.55–1.92 Mb in size. Array CGH plots for all 24 chromosomes are shown in the online Figure 5 (see the online Fig. 5 at http://www.interscience.wiley.com/jpages/1552-4825/suppmat/index.html). Both parents proved not to harbor the deletion as shown by array CGH.
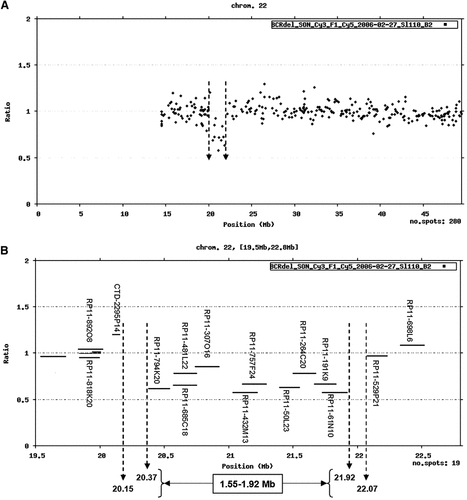
32K BAC Array CGH analysis of chromosome 22. A: Whole chromosome 22. B: Chromosome 22 from 19.5 to 22.8 Mb. Note the 1.55–1.92 Mb deletion (clones RP11-794K20 to RP11-61N10) indicated by the dashed arrows.
DISCUSSION
The clinical phenotypic description of microdeletion syndromes has usually preceded the understanding of the underlying causative cytogenetic rearrangements. This changed after the introduction of FISH-based screening tests, especially for subtle subtelomeric chromosomal rearrangements, which allowed the cytogenetic characterization of novel microdeletion syndromes in advance of their clinical recognition. With the introduction of array CGH technology, it is anticipated that more of these subtle chromosomal microdeletions and microduplications will be discovered and might prove to be more common than previously thought.
We report on a 15-year-old Hispanic boy with history of learning and behavioral problems. Physical examination showed normal growth with slight facial dysmorphism. A recent evaluation of our patient showed that his performance in the visual-motor integration test yielded an age equivalent of 8.4 years. FISH analysis clearly demonstrated that the patient carried a cryptic interstitial microdeletion on chromosome 22 band q11.2 that encompass the BCR gene but spares the DG/VCFs typically deleted region. Using a 32K BAC array CGH chip we were able to refine and precisely narrow the breakpoints. This microdeletion was estimated to be 1.55–1.92 Mb in size and to span approximately 20 genes. After performing a search on the “Human Genome Segmental Duplication Database” website (http://projects.tcag.ca/humandup/) it was found that this genomic region is flanked by LCR clusters that contain several modules; both in direct orientation as well as reverse orientation; with a very high degree of sequence homology (>95%) (Fig. 4), and ranging in size from ∼11 to ∼22 kb. The proximal LCR cluster flanking this microdeletion region represents the distal DG/VCFs LCR (LCR22-D). We are suggesting that these flanking LCRs could be implicated in the origin of our patient's microdeletion. The directly oriented homologous modules within the flanking LCR clusters could indeed mediate inter-chromosomal misalignment between the two chromosome 22 homologs during meiosis I and unequal cross-over leading to reciprocal deletion and duplication events. Reversely oriented modules on the other hand could be involved in intra-chromosomal recombination after formation of a “stem-loop” intermediate leading to a deletion event. Both inter- and intra-chromosomal recombination events have been reported for the typical 3.0 Mb deletion in DG/VCFs patients [Baumer et al., 1998; Edelmann et al., 1999]. Ravnan et al. reported eight other cases with the same interstitial deletion of the LSI BCR control FISH probe on 22q11.2 in a subtelomere FISH analysis of 11,688 cases with developmental disabilities. All eight patients had an intact DG/VCFs region as shown by FISH. The clinical phenotypes of these eight cases were discussed only briefly. Common features reported included developmental delay, hypotonia, and microcephaly [Ravnan et al., 2006]. Careful examination of the other LCR clusters telomeric to the DG/VCFs typically deleted region show that they also share a very high degree of sequence homology with each other and with the distal DG/VCFs LCR (LCR22-D), and therefore might mediate similar microdeletions and/or microduplications in the region just telomeric to the DG/VCFs typically deleted region (Fig. 4). The currently described microdeletion spanning the BCR gene (20.15–22.07 Mb) is located within the chromosomal segment previously indicated to contain the schwannomatosis predisposition gene [Jacoby et al., 1997; MacCollin et al., 2003]. It is probable that schwannomatosis is caused by mutations in a tumor suppressor gene, which has not yet been characterized. Assuming that the above hypothesis is correct, and also assuming that our microdeletion patient will not develop a schwannomatosis-related phenotype later in life, this 20.15–22.07 Mb interval on 22q is unlikely to contain the gene predisposing for the development of schwannomatosis.
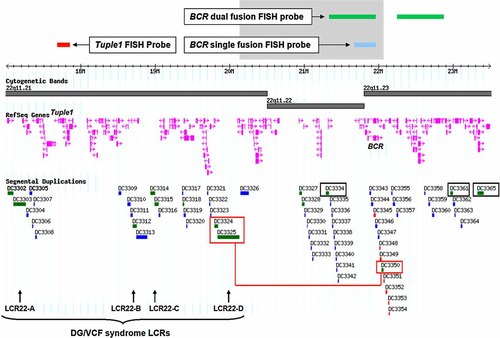
Genomic map of chromosome 22 from 17.0 to 23.5 Mb. FISH probes are illustrated as green, red and blue bars. The gray area represents our patient's deletion. The LCRs that are most likely implicated in the origin of this deletion are boxed in red. Note that our patient's microdeletion proximal flanking LCR represents the distal DG/VCFs LCR (LCR22-D). Other LCRs (black boxes) distal to the DG/VCFs critical region can also possibly mediate similar microdeletions and/or microduplications. Figure generated through “Human Genome Segmental Duplication Database” website. [Color figure can be viewed in the online issue, which is available at http://www.interscience.wiley.com.]
In conclusion, we suggest that our patient's microdeletion represents a previously unrecognized microdeletion syndrome on chromosome 22 just telomeric to the DG/VCFs typically deleted region and encompassing the BCR gene. Clinical features are subtle and include mild degree of mental impairment and mild facial dysmorphism, which necessitate testing using FISH and/or array CGH.
Acknowledgements
This study has been supported in part by the Swedish Cancer Society and the U.S. Army Medical Research and Materiel Command, award no. W81XWH-04-1-0269 to JPD.




