Angelman syndrome caused by an identical familial 1,487-kb deletion†
How to cite this article: Sato K, Iwakoshi M, Shimokawa O, Sakai H, Ohta T, Saitoh S, Miyake N, Niikawa N, Harada N, Saitsu H, Mizuguchi T, Matsumoto N. 2007. Angelman syndrome caused by an identical familial 1,487-kb deletion. Am J Med Genet Part A 143A:98–101.
To the Editor:
Angelman syndrome (AS, OMIM #105830) is a neurodevelopmental disorder characterized by mental retardation, ataxia, hypotonia, epilepsy, absence of speech, and specific facial features. At least four major mechanisms causing AS were validated: (i) an interstitial deletion of 15q11-q13 (70–75%), (ii) uniparental disomy (2–3%), (iii) imprinting defects (3–5%), (iv) UBE3A mutations (20%) [Clayton-Smith and Laan, 2003]. Most deletions are similar in size (approximate 4 Mb) and occur de novo through maternal unequal crossing over between low copy repeats (LCRs). Paternal occurrence of similar deletions, instead, results in Prader–Willi syndrome (PWS, OMIM #176270). In PWS no coding mutations have been found in contrast with UBE3A mutations in AS, suggesting that PWS is caused by loss of function of multiple genes.
Different sized deletions associated with AS are very rare. To our knowledge, at least three familial atypical deletions were reported [Saitoh et al., 1992; Buxton et al., 1994; Burger et al., 2002], but in one family [Buxton et al., 1994] a microdeletion could not be confirmed by another group [Sutcliffe et al., 1997]. In the remaining two families, microdeletions caused AS in maternal inheritance but no PWS features in paternal inheritance, enabling differentiation of the PWS critical region (PWSCR) from the AS critical region (ASCR).
We encountered a similar family with an atypical microdeletion through microarray CGH analysis of 30 individuals with idiopathic mental retardation [Miyake et al., 2006]. The family consisted of a boy, who was later confirmed with AS, and an asymptomatic mother and maternal grandfather (Fig. 1A). All three had an atypical microdeletion. Methylation PCR analysis [Kubota et al., 1997] of the proband showed a normal pattern (data not shown).
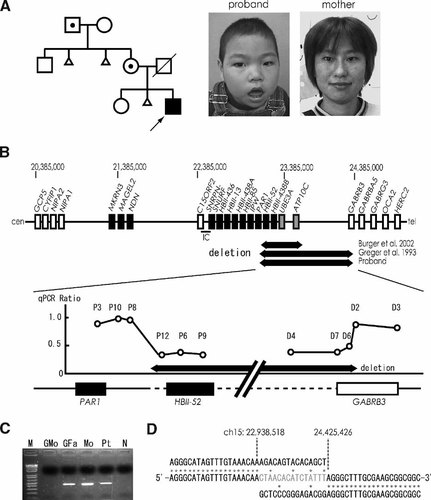
A: Pedigree of the family and photographs of the proband (arrow) and his mother. Proband, mother, and maternal grandfather (dotted) had a deletion. His father died of biliary atresia at 31 years old. B: Schematic presentation of genes (black boxes: genes expressed from paternal chromosome, gray boxes: genes expressed from maternal chromosome, open boxes: genes expressed from both chromosomes), rare familial deletions, and result of quantitative real-time PCR (qPCR) analysis. Deletion from HBII-52 cluster to GABRB3 was confirmed. C: Deletion specific PCR analysis of the family. PCR using primers, DSF and DSR, could successfully amplify a 2.6-kb product from grandfather (GFa), mother (Mo), and patient (Pt), but not from grandmother (GMo). M: size marker, N: negative control. D: Sequence of deletion breakpoints. The sequence in a middle row indicates the patient's sequence spanning the deletion. Upper and lower rows show normal sequences corresponding to centromeric and telomeric to the deletion. Proximal and distal deletion breakpoints are marked with UCSC coordinate chromosome 15 nucleotide positions. Fifteen nucleotides (in gray) were inserted. Asterisks indicate nucleotides identical to normal chromosome 15 sequence.
The deletion was intensively analyzed. FISH analysis using RPCI-11 BAC clones (701H24, 171C8, 1081A4, 607F22, 931B1, 2C7, 434O21, 203C13, 638J6, 899B22, 58D7, and 142M24) on the proband's metaphase chromosomes revealed that 1081A4, 607F22, 931B1, 2C7, 434O21, 203C13, 638J6, 899B22, and 58D7 were deleted, 171C8 and 142M24 were partially deleted, and 701H24 was not deleted (data not shown). Cosmid subclones constructed from BAC 171C8 were used for further FISH analysis. Cosmid D-2 was partially deleted (data not shown), indicating that the proximal deletion breakpoint was located in a region between UCSC coordinate chromosome 15 nucleotide 22,928,853 and 22,974,812 (end sequences of cosmid D-2). Subsequently quantitative real-time PCR (qPCR) using DyNAmo HS SYBR Green qPCR kit (Finnzymes, Espoo, Finland) was conducted to determine breakpoint locations according to the methods by Boehm et al. 2004 with some modification. A total of six sets of primers (P3, P10, P8, P12, P6, and P9) as test probes were designed within the cosmid D-2 region along with a control primer set for FBN1 locus at 15q21.1. Rotor-Gene™ 6200 HRM (Corbett Life Science, Sydney, Australia) could demonstrate a heterozygous deletion for primer-sets P12, P6, and P9 (Fig. 1B). Similarly five sets of primers (D4, D7, D6, D2, and D5) were selected from the region of BAC 142M24 and three sets, D4, D7, and D6, implied a deletion. Thus proximal and distal deletion breakpoints seemed to exist between P8 and P12 and between D6 and D2, respectively. Finally deletion breakpoints were successfully amplified as an approximate 2.6-kb product by PCR using LA-Taq (Takara Bio Inc., Otsu, Japan) and primers (DSF: 5′-TATAACTAGGTATTGGACTCATACTGAGGA-3′ and DSR: 5′-ACCTACAGCCTTCTAAGTACTGTATCCAT-3′) from the patient's DNA as well as his mother and maternal grandfather, but not from his maternal grandmother (Fig. 1C) or 10 normal controls. The PCR products were sequenced (Fig. 1D). The proband, his mother, and maternal grandfather had exactly the same breakpoint sequences. The deletion was 1,487-kb in size and contained HBII-52, HBII-438B, UBE3A, ATP10C, and a part of GABRB3 (Fig. 1B). Proximal and distal breakpoints were not related to any of PWS/AS-related LCRs [Christian et al., 1999]. It was surprising that the sequences were completely identical to those found in the family previously described [Greger et al., 1993]. Relationship of the two families could not be confirmed. However, coincidence of proximal and distal deletion breakpoints as well as inserted 15 nucleotides strongly suggests that the two families originated from the same ancestor. Both families indeed live in neighboring prefectures in Japan.
Phenotype of the proband is compatible to AS and was similar to that of the patient with the identical deletion described by Saitoh et al. 1992 (detailed clinical information was described by Sugimoto et al. 1992) (Table I). As his mother inherited the deletion from his maternal grandfather, she may suffer from some of PWS features if the deletion contains a gene(s) for PWS. She was carefully evaluated with regard to diagnostic criteria for PWS [Holm et al., 1993]. Only one major criterion (genetic microdeletion) and one minor (myopia) were recognized, thus PWS was definitely unlikely. This is consistent with the previous finding that a clinically healthy mother of three AS sibs, all sharing an identical deletion [Saitoh et al., 1992; Sugimoto et al., 1992].
| Patient | Proband | Patient 1a [Saitoh et al., 1992] |
B-5490 [Burger et al., 2002] |
|---|---|---|---|
| Psychomotor development | |||
| Mental retardation | Severe | Yes | Yes |
| Absence of speech | Yes | Yes | Yes |
| Able to speak single word | No | No | No |
| Able to make sound | Yes | ||
| Able to use sign language | Yes (only at urination) | ||
| Age walked alone | 2 years 7 months | 1 year 6 months | |
| Neurological features | |||
| Epilepsy | Yes | Yes | |
| EEG | Abnormal | Abnormal | No |
| Ataxia (When excited/running) (not marked) | Yes | Yes | Yes |
| Wide-based gait | Yes | ||
| Flapping hands (when excited/running) | Yes (slightly) | Yes | |
| Behavior | |||
| Happy disposition | Yes | ||
| Characteristic laughter | No | Yes | Yes |
| Physical features | |||
| Prominent mandible | Yes | Mild | |
| Small widely spaced teeth | Yes | Yes | |
| Large mouth | Yes | No | |
| Protruding tongue | Yes (slightly) | Yes | Yes |
| Small head (<25th percentile) | Yes | Yes | |
| Occiput | Normal | Flat | |
| Squint | Yes | Yes | |
| Weight | Within 90 percentile | −0.2 SD | |
| Height | Within 90 percentile | +0.8 SD | |
| Hypopigmentation | No | No | No |
| Miscellaneous | |||
| Sleep | Disrupted | ||
| Mouthing | No | ||
| Dribbling/drooling | Yes | ||
| Other | Father is dead (Biliary atresia) | ||
Recently brain-specific snoRNA HBII-52 was found to regulate alternative splicing of serotonin receptor 2C, possibly influencing the serotonine response [Kishore and Stamm, 2006]. Clustering 47 copies of HBII-52 are maternally imprinted and are suggested to play an important role in pathogenesis of PWS [Cavaille et al., 2000]. However in this family as well as the previously described family [Saitoh et al., 1992], two healthy mothers possessed the microdeletion inherited from their fathers and the deletion included the complete HBII-52 locus. Thus, the normal phenotype of the mother described here again supports that HBII-52 does not play any roles in PWS. Similarly the paternally inherited 570-kb deletion in a healthy mother also included HBII-52 [Burger et al., 2002; Runte et al., 2005]. Rare balanced translocations involving paternal 15q11-q13 in PWS and rare atypical microdeletions now delimit the PWSCR to a 121-kb region covering HBII-438A and HBII-85 clusters [Wirth et al., 2001; Gallagher et al., 2002].
In conclusion, a very rare identical 1,487-kb deletion was found in two families possibly originating from the same ancestor. It should be stressed that the deletion can be inherited without any symptoms through paternal lines. Finally HBII-52 may not be important for PWS pathogenesis.
Acknowledgements
Research grant from the Ministry of Health, Labour and Welfare for N.M., Grant-in-Aid for Scientific Research on Priority Areas (Research on Pathomechanisms of Brain Disorders) for N.M., Research Promotion Fund from Yokohama Foundation for Advancement of Medical Science for N.M., Natural Science Research Fund from the Mitsubishi Foundation for N.M., and SORST from Japan Science and Technology Agency (JST) for N.N.




