3′ UTR polymorphism of the serotonin transporter gene and sudden infant death syndrome: Haplotype analysis†
How to cite this article: Maher BS, Marazita ML, Rand C, Zhou L, Berry-Kravis EM, Weese-Mayer DE. 2006. 3′ UTR polymorphism of the serotonin transporter gene and sudden infant death syndrome: Haplotype analysis. Am J Med Genet Part A 140A:1453–1457.
To the Editor:
The precise etiology of sudden infant death syndrome (SIDS) remains to be elucidated, but much work has been done to understand risk factors, both biological and environmental. Among these risk factors, the role of genes in several candidate risk pathways has been elucidated. For example, some studies implicate cardiac sodium channel genes (SCN5A) with a role in cardiac events during sleep leading to SIDS [Schwartz et al., 1998, 2000; Ackerman et al., 2001; Plant et al., 2006]. Other reports implicate genes pertinent to the early embryologic origin of the autonomic nervous system (ANS) [Weese-Mayer et al., 2004], a system thought to be malregulated in SIDS [Kelly et al., 1986; Schechtman et al., 1988; Kahn et al., 1992; Ponsonby et al., 1992; Meny et al., 1994; Franco et al., 1998, 1999; Ledwidge et al., 1998; Schwartz et al., 1998].
Serotonin pathways have been of particular interest in SIDS because serotonin influences a broad range of physiological systems including the regulation of breathing, the cardiovascular system, temperature, and the sleep–wake cycle. Panigrahy et al. 2000 reported a decrease in serotonergic receptor binding in the arcuate nucleus, n. raphé obscurus, and other medullary regions that contain serotonergic cell bodies in SIDS cases in the US. Similarly, Ozawa and Okado 2002 reported a decrease in serotonergic receptor binding in the dorsal nucleus of the vagus, solitary nucleus and ventrolateral medulla in SIDS cases in Japan.
Although the serotonergic system is complex, the serotonin transporter (5HTT) is a major molecular determinant of serotonergic function. Two functional polymorphisms in the serotonin transporter gene (5HTT) are known to modify gene transcription and transporter expression. These include a promoter insertion/deletion polymorphism [Heils et al., 1996] and a variable number tandem repeat (VNTR) in intron 2 [Fiskerstrand et al., 1999]. The long allele of the promoter polymorphism and the 12 repeat allele of the intron 2 VNTR are both associated with enhanced promoter function, increased 5HTT transcription, increased transporter protein expression, and presumably increased cellular 5HT uptake and lower synaptic levels of 5HT.
Narita et al. 2001 initially identified an association between the long allele (L) of the 5HTT promoter polymorphism and SIDS in a small Japanese cohort. Weese-Mayer et al. 2003a replicated this association finding in a larger sample of Caucasian and African-American SIDS cases and gender/ethnicity-matched controls. A notable finding was the increased frequency of the long allele in African-American populations as contrasted with the decreased frequency in the Japanese population, both consistent with the patterns of SIDS prevalence in those ethnic populations. In a subsequent study, Weese-Mayer et al. 2003b found an association between SIDS and the 12 repeat allele of the intron 2 VNTR and the L-12 haplotype (“long” allele present at the promoter and “12” allele present at intron 2 on the same chromosome) in the African-American subgroup but not in Caucasians. These studies provide strong evidence that genetic variation in 5HTT contributes to SIDS susceptibility, perhaps through a mechanism involving lower extraneuronal 5HT levels during development.
Although two functional polymorphisms in 5HTT have been shown to be associated with SIDS, a full assessment of the contribution of genetic variation in 5HTT to SIDS risk requires assessment of multiple markers in the vicinity of the gene. In this report, we examine the association between SIDS and a polymorphism in a putative polyadenylation site in the 3′ UTR of 5HTT [Battersby et al., 1999] and between SIDS and 5HTT haplotypes including the 3′ UTR SNP, intron 2 VNTR and the promoter polymorphism, in an effort to further clarify the relationship between allelic variation in 5HTT and SIDS.
Two distinct groups were investigated in this study: 92 SIDS cases and 92 gender- and ethnicity-matched control subjects, which were included in previous publications [Weese-Mayer et al., 2003a,b, 2004]. SIDS cases (mean age at death 96 days ± 52) with a diagnosis made by the University of Maryland Medical Examiner were identified in the University of Maryland Brain & Tissue Bank (http://medschool.umaryland.edu/btbank/main.html) (21 African-American females, 25 African-American males, 16 Caucasian females, and 30 Caucasian males). The diagnosis of SIDS was based on the accepted definition [Willinger et al., 1991]. A three-generation family history was taken for each control to ensure that no family member had a diagnosis of SIDS, Hirschsprung disease, Congenital Central Hypoventilation Syndrome, apparent life threatening event, primary disorder of ANS dysregulation, or tumor of neural crest origin. To ensure that the controls were not likely to succumb to SIDS, the control subjects were always older than 1 year of age. This study was approved by the Rush University Medical Center and the University of Pittsburgh institutional review boards, and informed consent was obtained from all control subjects.
Genomic DNA was isolated from frozen frontal cortex brain tissue (SIDS cases) and blood (controls) samples utilizing standard methods [Weese-Mayer et al., 2003a]. Target DNA containing the 3′ UTR polymorphism was amplified using primer pair: HTT.PCR1: 5′-CCGCTTGAATGCTGTGTAACACAC-3′; and HTT.PCR3: 5′-GTACCCTTCCAATAATAACCTCC-3′ as previously described [Battersby et al., 1999]. PCR products (17 µl) were digested overnight with Tru9 1 (Promega, Fitchburg, WI) according to the manufacturer's protocol and visualized on 2% agarose gel using ethidium bromide [Battersby et al., 1999]. The 5HTT promoter polymorphism and intron 2 polymorphism were genotyped as reported previously [Weese-Mayer et al., 2003a,b].
For each case-control comparison we computed standard χ2 tests of independence between genotype or allele frequency distribution and SIDS case/control status. Under the null hypothesis of no genotype distribution or allele frequency differences, the test statistic follows a χ2 distribution with degrees of freedom equal to one less than the number of genotypes or alleles. Haplotype frequencies were estimated using a statistical approach to reconstruct haplotypes from genotype data [Stephens et al., 2001] using 10,000 iterations. Estimation of haplotype frequencies was conducted separately for Caucasian and African-American samples. Statistical significance was assessed via a “Product of Approximated Conditionals” method [Li and Stephens, 2003]. Standard measures of linkage disequilibrium (D′) were calculated for each ethnic group.
Analyses were performed for genotype distribution and allele frequency comparing the 92 SIDS cases and 92 screened ethnicity- and gender-matched control subjects. Data are provided in Table I. The genotype distributions for the 3′ UTR polymorphism did not differ significantly between the SIDS cases and matched controls over the total dataset (92 matched pairs; χ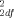 = 0.43, P = 0.81) (Table I), between the Caucasian SIDS cases and controls (46 matched pairs: χ
= 0.43, P = 0.81) (Table I), between the Caucasian SIDS cases and controls (46 matched pairs: χ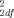 = 3.25, P = 0.20), nor between the African-American SIDS cases and controls (46 matched pairs; χ
= 3.25, P = 0.20), nor between the African-American SIDS cases and controls (46 matched pairs; χ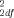 = 0.85, P = 0.65). The allele frequencies for the 3′ UTR polymorphism did not differ significantly between the SIDS cases and matched controls over the total dataset (92 matched pairs; χ
= 0.85, P = 0.65). The allele frequencies for the 3′ UTR polymorphism did not differ significantly between the SIDS cases and matched controls over the total dataset (92 matched pairs; χ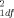 = 0.11, P = 0.74) (Table I), between the Caucasian SIDS cases and controls (46 matched pairs: χ
= 0.11, P = 0.74) (Table I), between the Caucasian SIDS cases and controls (46 matched pairs: χ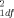 = 0.83, P = 0.36), nor between the African-American SIDS cases and controls (46 matched pairs; χ
= 0.83, P = 0.36), nor between the African-American SIDS cases and controls (46 matched pairs; χ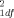 = 0.12, P = 0.73).
= 0.12, P = 0.73).
| Intron 2 | Control | SIDS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Caucasian | African-American | Total | Caucasian | African-American | Total | |||||||
| Genotype | n | % | n | % | n | % | n | % | n | % | n | % |
| G/G | 9 | 19.56 | 5 | 10.87 | 14 | 15.22 | 10 | 21.74 | 4 | 8.70 | 14 | 15.22 |
| G/T | 20 | 43.48 | 12 | 26.09 | 32 | 34.78 | 12 | 26.09 | 16 | 34.78 | 28 | 30.43 |
| T/T | 17 | 36.96 | 29 | 63.04 | 46 | 50.00 | 24 | 52.17 | 26 | 56.52 | 50 | 54.35 |
| Allele | ||||||||||||
| G | 38 | 41.30 | 22 | 23.91 | 60 | 32.61 | 32 | 34.78 | 24 | 26.09 | 56 | 30.43 |
| T | 54 | 58.70 | 70 | 76.09 | 124 | 67.39 | 60 | 65.22 | 68 | 73.91 | 128 | 69.57 |
- a Please refer to text for statistical analysis of total dataset and inter-group comparisons.
Analyses were performed for haplotypes spanning the 5HTT gene (promoter, intron 2 and 3′ UTR variants) for the total dataset and for the Caucasian and African-American subgroups. The 3′ UTR genotypes were obtained in the same SIDS-control pairs as the promoter and intron 2 genotypes [Weese-Mayer et al., 2003a,b]. Data for all three-marker genotype combinations are presented in Table II.
| Haplotypeb | Control | SIDS | ||||||
|---|---|---|---|---|---|---|---|---|
| Promoter | Intron 2 | 3′ UTR | Caucasian | African-American | Total | Caucasian | African-American | Total |
| S | 9 & 10 | G | 0.003 (0.006) | 0.019 (0.009) | 0.018 (0.007) | 0.004 (0.008) | 0.003 (0.005) | 0.008 (0.006) |
| S | 9 & 10 | T | 0.168 (0.016) | 0.034 (0.015) | 0.100 (0.012) | 0.070 (0.015) | 0.025 (0.009) | 0.049 (0.011) |
| S | 12 | G | 0.280 (0.016) | 0.071 (0.011) | 0.173 (0.011) | 0.182 (0.016) | 0.078 (0.014) | 0.129 (0.011) |
| S | 12 | T | 0.081 (0.014) | 0.162 (0.016) | 0.124 (0.011) | 0.102 (0.012) | 0.092 (0.016) | 0.092 (0.010) |
| L | 9 & 10 | G | 0.016 (0.010) | 0.024 (0.010) | 0.018 (0.008) | 0.022 (0.010) | 0.023 (0.012) | 0.017 (0.009) |
| L | 9 & 10 | T | 0.237 (0.019) | 0.186 (0.018) | 0.216 (0.015) | 0.291 (0.016) | 0.092 (0.015) | 0.196 (0.013) |
| L | 12 | G | 0.114 (0.017) | 0.125 (0.011) | 0.118 (0.012) | 0.138 (0.016) | 0.156 (0.016) | 0.150 (0.012) |
| L | 12 | T | 0.101 (0.016) | 0.378 (0.018) | 0.234 (0.014) | 0.189 (0.013) | 0.530 (0.019) | 0.358 (0.012) |
The estimated haplotype frequencies (Table II) were used to calculate linkage disequilibrium (LD) measures in the total dataset and for the Caucasian and African-American subgroups. LD between the 3′ UTR and the promoter and intron 2 polymorphisms was highly significant in both the Caucasian (3′ UTR and promoter: D′ = 0.41, χ2 = 23.31, P < 0.0001; 3′ UTR and intron 2: D′ = 0.90, χ2 = 62.25, P < 0.0001) samples and the African-American (3′ UTR and promoter: D′ = 0.273, χ2 = 12.90, P = 0.0003; 3′ UTR and intron 2: D′ = 0.676, χ2 = 7.05, P = 0.008) samples. However, for the total dataset and each of the ethnic subgroups there were no significant differences in haplotype frequency distribution (combined: P = 0.06; African-American: P = 0.13; Caucasian: P = 0.11).
Our group previously reported significant association between SIDS and polymorphisms in the serotonin transporter promoter region as well as the intron 2 polymorphism [Weese-Mayer et al., 2003a,b]. In this report, the analysis of the impact of genetic variation in 5HTT on SIDS risk was extended to a 3′ UTR polymorphism. We found no association between SIDS and the 3′ UTR SNP in either ethnic group studied. Moreover, inclusion of the 3′ UTR SNP in haplotype association analyses decreased the significance of the previously observed 5HTT associations.
The 3′ UTR SNP is a polymorphism in 5HTT identified by Battersby et al. 1999 and located within a putative polyadenylation signal for one of the commonly used polyadenylation sites for the 5HTT mRNA. Although allelic variation at the site was not shown to substantially influence polyadenylation site usage [Battersby et al., 1999], the 3′RACE assay used to assess polyadenylation was not quantitative and it remains possible that minor abnormalities in polyadenylation in vivo might affect stability of 5HTT mRNA and/or transport into the cytoplasm. A subsequent study [Melke et al., 2003] was unable to identify any effect of this 3′ UTR polymorphism on the platelet serotonin transporter expression assayed by [3H]paroxetine binding. Likewise, no association has been detected between the 3′ UTR polymorphism and major depression or bipolar affective disorder [Battersby et al., 1999], or premenstrual dysphoria [Melke et al., 2003]. These data suggest that the 3′ UTR may not really be a functional polymorphism, although it is still expected to be a useful tool for assessment of the contribution of genetic variation in the serotonin transporter gene to disease. Similar to the findings with the SIDS cohort presented here, the 5HTT promoter polymorphism was recently shown to be associated with attention deficit hyperactivity disorder, while the 3′ UTR was not associated [Curran et al., 2005]. Findings in both studies likely relate to the difference in functional effects of the promoter polymorphism and the 3′ UTR SNP.
Although genetic findings for any disease are intriguing, it is important to consider all sources of risk. In fact, many environmental factors including parental smoking [Schoendorf and Kiely, 1992], socio-economic status [Taylor and Sanderson, 1995], sleeping position [Fleming et al., 1990; Dwyer et al., 1991a,b; Mitchell et al., 1992], and infection [Blackwell et al., 2004] have been identified as risk factors for SIDS. Recently, several authors have posited that interaction between genes and environment may be an important risk factor in SIDS etiology [Opdal and Rognum, 2004; Hunt, 2005; Plant et al., 2006]. Interestingly, the 5HTT promoter polymorphism has previously been implicated in gene-environment interactions [Caspi et al., 2003]. Moreover, the mechanism for this interaction may have already been identified. That is, variation in the promoter polymorphism mediates serotonin transporter expression in response to glucocorticoid [Glatz et al., 2003]. Glatz et al. 2003 propose that variation in the promoter polymorphism may influence individual risk for diseases related to stress response. Therefore, based on the findings from this and previous studies, it appears that the influence of polymorphisms in 5HTT on SIDS risk may be mediated by specific effects on promoter activity and responsiveness, transporter expression and resultant modulation of 5HT levels in the CNS, such that SIDS risk occurs in association with increased transporter and lower extraneuronal 5HT levels. Under this hypothesis, the 3′ UTR SNP would not be predicted to directly influence SIDS risk, as this polymorphism does not appear to affect transporter expression. Future investigations on the influence of 5HTT on SIDS risk should focus on polymorphisms which directly impact regulation of transporter protein expression or function.
Acknowledgements
We thank the parents of the SIDS cases for their gracious tissue donations, as well as the control subjects who shared their DNA. Sources of support: C.J. Foundation for SIDS, the Justin Carl Suth SIDS Research Fund, the Joseph Tyler Gertler SIDS Research Fund, and The University of Maryland Brain & Tissue Bank for Developmental Disorders (D.E. W.-M.); Spastic Paralysis and Allied Diseases of the Central Nervous System Research Foundation of the Illinois-Eastern Iowa District Kiwanis International (E.M.B.-K.).




