Probable identity-by-descent for a mutation in the Dyggve–Melchior–Clausen/Smith–McCort dysplasia (Dymeclin) gene among patients from Guam, Chile, Argentina, and Spain
To the Editor:
Dyggve–Melchior–Clausen dysplasia (DMC; OMIM 223800) [Dyggve et al., 1962] and Smith–McCort dysplasia (SMC; OMIM 607326) [Smith and McCort, 1958] are allelic, recessively-inherited osteochondrodysplasia phenotypes. Clinical features common to both conditions include short limbs and short trunk, a barrel shaped chest, kyphoscoliosis, microcephaly and brachydactyly. The two disorders are distinguished by the presence of mental retardation in DMC patients. Radiographically, patients with DMC and SMC exhibit platyspondyly, a characteristic double-humped appearance of the vertebrae with a central constriction, and both metaphyseal and epiphyseal irregularities [Burns et al., 2003]. The phenotypes thus fall within the spondyloepimetaphyseal dysplasia subgroup of the osteochondrodysplasias [Hall, 2002]. A lacy appearance of the iliac crests is a pathognomonic finding. Growth plate histology shows sparse chondrocytes, with disorganization and lack of column formation in the hypertrophic zone. Electron microscopy reveals that chondrocytes have dilated endoplasmic reticulum containing amorphous material of unknown constitution [Nakamura et al., 1997].
DMC and SMC are caused by mutations in the FLJ20071/DYMECLIN gene [Cohn et al., 2003; El Ghouzzi et al., 2003]. Homozygosity or compound heterozygosity for null mutations was shown to be the primary cause of DMC, while two SMC patients from Guam were compound heterozygtes for an exon-skipping mutation and a missense mutation [Cohn et al., 2003]. The exon-skipping mutation was an A > G transition in the splice acceptor of intron 7 (IVS7-2A > G), producing a transcript lacking exon 8. This resulted in an altered reading frame, predicting premature termination of protein synthesis. The same mutation was recently described in a Spanish DMC patient who was a compound heterozygote for this and another splicing mutation [Paupe et al., 2004].
We studied three DMC families, one each from Chile, Argentina, and Spain. Clinical information, radiographs, and blood samples were obtained under an IRB approved protocol. The clinical phenotypes of the two SMC patients from Guam (International Skeletal Dysplasia Registry reference numbers R94-126 and R98-116) were described previously [Cohn et al., 2003].
The DMC patient from Chile (R03-256), was a female born to consanguineous parents by C-section at 34 weeks gestation due to preeclampsia, and had a birth weight of 2,460 g (10th–25th centile), and length of 45 cm (25th–50th centile). She manifested early onset growth retardation and bilateral hip subluxations. At 11 years of age her height was 84 cm (below 3rd centile), she had coarse facial features with prominent supraciliary arches, a short thorax with prominent ribs and sternum, coxa vara, genu valgum, and prominent wrists and ankles. She had severe mental retardation, with absent speech, communicating by whistling or hissing. She was ambulatory but required assistance for most activities of daily living.
The DMC patient from Argentina (R03-318), was a male born at term to a consanguineous couple of Spanish descent. Birth weight, length, and head circumference were normal, but soon hypotonia, psychomotor delay, slow growth, and sternal prominence ensued. A skeletal survey at age 10 months led to the diagnosis. At 6 years, 7 months, he had a waddling gait, and was short, slim, borderline microcephalic (2nd centile), and mentally retarded; he also had a long and narrow face, bilateral superior epicanthal folds, small teeth, bifid uvula, prognathism, a short neck and trunk, prominent hips, dorsal-lumbar kyphosis, short limbs (especially the proximal segments), small hands and feet, and limited extension of the elbows and knees. Skeletal features apparent on radiographs included advanced bone age, dorsal-lumbar kyphosis, platyspondyly with notched upper and lower vertebral body surfaces, small iliac bones with irregularly calcified borders, subluxated hips, short metacarpals and phalanges, an accessory distal epiphysis on the 1st metacarpal, and cone-shaped distal epiphyses in all phalanges.
The DMC patient from Spain (R04-573), was the first child of a non-consanguineous couple. The delivery occurred spontaneously at 39 weeks gestation, and the birth weight was 2,990 g (25th–50th centile), length 48 cm (50th centile), and occipito-frontal-circumference 33 cm (50th centile). Clinical examination at birth was unremarkable apart from sternal bulging. At 3 months of life, a thoracic deformation was noted, and X-rays showed platyspondyly. At 8 years of age the patient presented a coarse face with a bulky jaw, a barrel-shaped chest, and clear short trunk dwarfism. At this time, he had mild to moderate mental retardation. There was progressive genu valgum, subsequently surgically corrected at 9 years of age, thoracic kyphosis, and lumbar lordosis. The radiographic findings included platyspondyly with markedly flattened vertebral bodies, a lacy appearance of the iliac crests, dysplastic acetabulae, short long bones and short hands.
Mutation analysis was carried out by amplification and sequence analysis of the 17 coding exons and flanking splice junction consensus sequences of the DYMECLIN gene, as described previously [Cohn et al., 2003; El Ghouzzi et al., 2003]. The Chilean and Argentine patients, both the offspring of consanguineous parents, were found to be homozygous for a mutation in the intron 7 splice acceptor (IVS7-2A > G), predicting skipping of exon 8, yielding a transcript with a frameshift and premature termination at codon 269. This mutation was also identified in the Spanish DMC patient, who was a compound heterozygote with a second mutation occurring in the splice acceptor of intron 5 (IVS5-2A > G), predicting skipping of exon 6. The genotype of this patient was the same as that of an independent Spanish DMC case [Paupe et al., 2004]. The IVS7-2A > G mutation was previously identified in two independent SMC families from Guam [Cohn et al., 2003] in which the affected individuals were compound heterozygotes for IVS7-2A > G, and a missense mutation predicting an E87K substitution (cDNA change: 259G > A). Analysis of mRNA from cartilage in the patients from Guam, demonstrated that the IVS7-2A > G mutation resulted in leaky skipping of exon 8 [Cohn et al., 2003]. The presence of a small amount of normally spliced mRNA, which would be predicted to lead to some normal gene product, was posited as an explanation for the clinically milder SMC phenotype seen in the patients. The data presented here disprove that hypothesis by demonstrating that homozygosity for the mutation produces DMC. Although the possibility of modifier loci contributing to phenotypic expression cannot be ruled out, identification of homozygosity for the mutation in two independent patients suggests that such hypothetical modifiers would have to be reasonably frequent in the populations from which the patients originated. We thus favor the hypothesis that homozygosity for the mutation alone is sufficient to produce DMC.
To investigate whether finding the IVS7-2A > G mutations in the five apparently unrelated families reflects identity by descent or a mutational hotspot, genotypes were determined at ten intragenic single nucleotide polymorphisms (SNPs) located across the length of the gene (Fig. 1a). SNPs were identified within the FLJ20071/DYMECLIN locus using dbSNP [http://www.ncbi.nlm.nih.gov/SNP/]. Ten SNPs, with heterozygosities ranging from 0.3 to 0.47 were selected. The SNP loci were amplified and sequenced using PCR primers designed using Primer 3 software [http://frodo.wi.mit.edu/primer3/primer3_code.html]. SNP haplotypes were derived by analysis of allele segregation among family members. The SNPs were selected to flank both the IVS7-2A > G mutation (between rs3809924 and rs530550) and the 259G > A mutation (between rs4939867 and rs3809924).
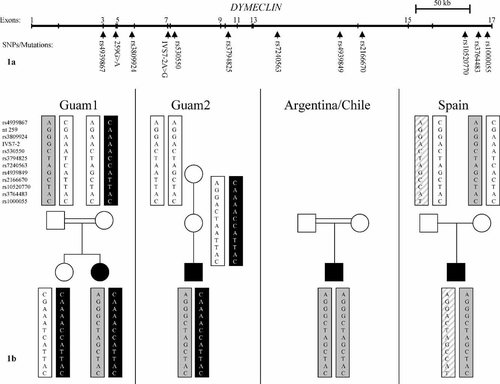
a: Schematic representation of the DYMECLIN gene with the positions of the 10 SNPs used in haplotype analysis shown below. Vertical lines represent exons, which are numbered above. Also indicated are the relative positions of the IVS7-2A > G and 259G > A mutations. The entire gene spans 420 kb of genomic DNA. b: Pedigrees showing SNP haplotypes associated with the IVS7-2A > G (gray box), 259G > A (black box, white text), IVS5-2A > G (gray hatched box), and wild type (white box) Dymeclin alleles in the five families. SNPs are listed according to their “rs” numbers to the left of the paternal haplotypes in family Guam1. Also included are genotypes at the nucleotides mutated in the Guam families; nt 259 refers to the cDNA position of the 259G > A mutation predicting the E87K substitution; IVS7-2 refers to the genomic DNA position of the IVS7-2A > G mutation predicting skipping of exon 8.
For the two SMC patients from Guam and the Spanish DMC patient, examination of the parental and/or grandparental SNP alleles in each family, together with the known parental origin of the mutations [Cohn et al., 2003; Paupe et al., 2004], allowed determination of the haplotype associated with each mutation (Fig. 1b). However, for the affected individual in family Guam 2, lack of DNA from the father made it necessary to assume that no intragenic recombination had occurred. For the 259G > A allele, SNP analysis showed a single haplotype was associated with the mutation in the Guam families (Fig. 1b). These data suggest that this mutation arose in a common ancestor.
A single haplotype was also associated with the IVS7-2A > G mutation in the two SMC families from Guam (Fig. 1). These data suggest that this mutation in these patients also arose in a common ancestor. The haplotypes of the two South American patients showed that both were homozygous for a single haplotype that was identical to the haplotype associated with the IVS7-2A > G mutation in the Guam families. The same haplotype was also shown to co-segregate with the IVS7-2A > G mutation in the Spanish patient. This suggests the hypothesis that the IVS7-2A > G mutation is identical by descent among all of these populations. Absence of haplotype data for ethnically matched individuals from the four countries involved limits the ability to make this conclusion from current data [Nolte and Te Meerman, 2002]. However it has been argued that coincidental haplotype sharing concordant with the segregation of the same mutation in a number of families is highly improbable, thereby implying that identity by descent is more likely [Morar et al., 2004].
The possibility of identity by descent is supported by examination of historical data regarding the colonization of Argentina, Chile, and Guam by European explorers from the 16th century onward. The Portuguese explorer Ferdinand Magellan sailed from Spain in the year 1519, with the intention of completing the first circumnavigation of the Earth. On his voyage, he visited, among other places, Argentina, Chile, and Guam [http://campus.northpark.edu/history/WebChron/WestEurope/Magellan.html]. Further Spanish explorers spent the remainder of the 16th century populating each of these countries, and each of them retains a strong Spanish influence to this day. This suggests the possibility that the exon-skipping mutation and haplotype referred to here originated in Spain or a closely related population and, sometime during the past 500 years, was carried around the world by travelers.
Numerous studies have used combined mutation and haplotype data to suggest founder effects that are consistent with the known migration patterns of the populations involved [Uyguner et al., 2003; Lucas et al., 2005; Najmabadi et al., 2005; Rodriguez et al., 2005]. Others have tracked the origins of mutations based on frequency gradients of the associated haplotypes [Levo et al., 1999; Lucotte, 2001], or used common haplotypes to suggest relationships among families [Le Bizec et al., 2005]. These models imply that rare mutations mark specific haplotypes, thereby allowing historical population migrations to be traced. For IVS7-2A > G, observed in the DMC and SMC patients studied herein, we suggest that presence of the mutation on the same haplotype in patients from diverse geographic regions define historical genetic relationships that reflect the colonial migrations contributing to the settlement of the New World.




