A novel missense mutation in a C2 domain of OTOF results in autosomal recessive auditory neuropathy
Abstract
Screening of 12 Turkish families with apparently autosomal recessive nonsyndromic sensorineural deafness without GJB2 and mtDNA m.1555A > G mutations for 11 previously mapped recessive deafness loci showed a family in which hearing loss cosegregated with the DFNB9 (OTOF) locus. Three affected children were later found to carry a novel homozygous c.3032T > C (p.Leu1011Pro) mutation in the OTOF gene. Both parents were heterozygous for the mutation. p.Leu1011Pro alters a conserved leucine residue in the C2D domain of otoferlin. Pure tone audiometry of the family showed severe to profound sensorineural hearing loss (with U-shape audiograms) in children, and normal hearing in the parents. Otoacoustic emissions and auditory brainstem response (ABR) suggested the presence of auditory neuropathy in affected individuals. © 2005 Wiley-Liss, Inc.
INTRODUCTION
Congenital or prelingual-onset deafness affects 1 in 800 newborns, of whom half are thought to be caused by genetic factors. Mutations in a number of genes with autosomal recessive inheritance are responsible for approximately 80% of genetic cases [Morton, 1991; Marazita et al., 1993]. A total of 53 different loci and 20 different genes are listed for autosomal recessive hearing impairment on the hereditary hearing loss homepage (http://webhost.ua.ac.be/hhh/), demonstrating genetic heterogeneity for this most common type of sensorial disorder.
Mutations in GJB2, encoding connexin 26, have been reported as the most common cause of autosomal recessive deafness, making up to 50% of all recessive families in some populations [Kenneson et al., 2002]. Each of the remaining identified genes, however, has been demonstrated to be responsible for deafness in single or few families. Mutations in SLC26A4 are exceptional examples where a considerable number of carrier families from different parts of the world have been reported [Park et al., 2003; Blons et al., 2004].
In this study, we report on the results of screening for 11 known recessive loci and genes in families with presumed autosomal recessive deafness from Turkey.
MATERIALS AND METHODS
The study was approved by the Institutional Ethics Committee of Ankara University (#30817 on 11/14/2001). Each participant or parent signed an informed consent prior to participation in the study.
A total of 12 families, in which at least two siblings had congenital or prelingual-onset severe to profound sensorineural hearing loss with normal hearing parents, were included in the study. All families were originating from small towns or villages in different parts of Anatolia. Parental consanguinity was present in 10 families. All families were evaluated and found to have no syndromic findings or a history of environmental exposure. All probands were evaluated with a standard pure tone audiometry, speech discrimination, acoustic middle ear muscle reflexes and additional auditory brainstem response (ABR), and/or otoacoustic emissions, where necessary.
DNA samples of all participants were obtained from peripheral blood using a standard phenol chloroform method. All samples were initially screened and found to be negative for the GJB2 mutations, Δ(GJB6-D13S1830) deletion, and mitochondrial m.1555A > G mutation.
The families were later screened for cosegregation of deafness and genotypes of microsatellite markers for DFNB1 (GJB2 and GJB6), DFNB2 (MYO7A), DFNB3 (MYO15), DFNB4 (SLC26A4), DFNB7(TMC1), DFNB8 (TMPRSS3), DFNB9 (OTOF), DFNB12 (CDH23), DFNB16 (STRC), DFNB21 (TECTA), and DFNB30 (MYO3A). Microsatellite markers listed on the hereditary hearing loss homepage (http://webhost.ua.ac.be/hhh/) were selected with help of relevant references and used during these studies. Following standard PCR reactions, PCR products were denatured, loaded on 6% denaturing polyacrylamide gels, and run on a vertical polyacrylamide gel system (Life Technologies Model S2001, Gibco BRL, Gaithersburg, MD, USA). Silver staining was used for visualization. Two CEPH DNA samples were simultaneously run for sizing.
The coding exons and surrounding regions of OTOF were screened in two families, where cosegregation of deafness with microsatellite alleles was evident, using PCR-SSCP protocols [Mirghomizadeh et al., 2002; primer sequences used in these studies are available on request]. The PCR products were denatured at 95°C, loaded on 7% non-denaturing polyacrylamide gels, run overnight at +4°C with a vertical gel electrophoresis system (Protean II Xi Cell, BioRad, Hercules, CA, USA), and visualized with silver staining. Samples showing band changes were directly sequenced with an automated sequencer (Beckman Coulter CEQ 2000XL) following cycle sequencing reactions and analyzed using Beckman Coulter Software (version 1.1).
A sequence change (c.3032T > C), found in OTOF in one family, was screened with PCR-RFLP using MspI restriction enzyme (Fermentas, Vilnius, Lithuania) in 50 unrelated hearing individuals and 50 deaf probands from Turkey. Genomic DNA was amplified using forward 5′ GTT CCG TGG AAG CTG AGT G 3′ and reverse 5′ TCA GCG CAG GTG GAG TGC 3′ primers. When the 227 bp PCR product was digested with MspI, two bands, at 164 bp and 63 bp, were present in homozygote mutants since the enzyme recognizes the mutant allele. Heterozygotes showed three bands at 227 bp, 164 bp, and 63 bp. Wild type alleles showed only one band at 227 bp.
RESULTS
Cosegregation of microsatellite alleles for DFNB9 (OTOF) with deafness was observed in two families. None of the other loci were found to be segregating with deafness in the remaining families. Further screening of OTOF resulted in identification of a novel missense change, c.3032T > C (p.Leu1011Pro), in exon 26 in one family (according to exon numbering by Yasunaga et al., 2000; gene bank accession number nm_194248 for transcript variant 1 encoding isoform a)(Figs. 1 and 2). The parents, who were second cousins, were heterozygous, and their three deaf children were homozygous for this alteration. Screening for this change in 50 deaf and 50 hearing individuals was negative. No mutation was found in the second family suggesting linkage to the DFNB9 locus.
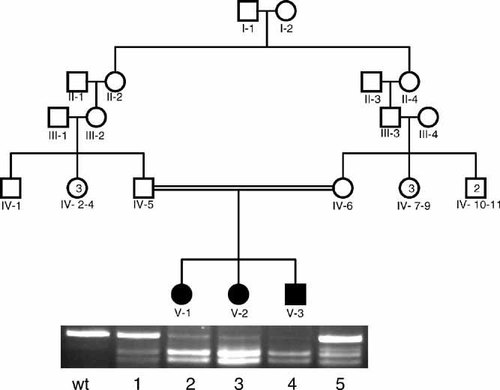
Pedigree of the family with the c.3032T > C mutation in OTOF. Results of screening for c.3032T > C with MspI are shown in a gel picture. Wt: Wild type; lanes 1, 5 are heterozygous father and mother, respectively, showing three bands at 227 bp, 164 bp, and 63 bp; lanes 2–4 are homozygous children showing two bands at 164 bp and 63 bp.
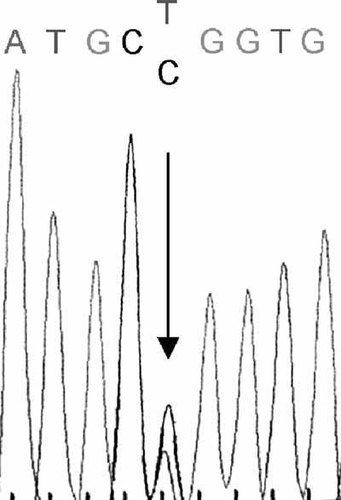
Heterozygous c.3032T > C mutation in OTOF.
Audiological investigation in the family showed that three children had severe to profound congenital or prelingual onset sensorineural hearing loss (Fig. 3). They did not have any predisposing factors such as hyperbilirubinemia or prematurity in the medical history and their family history was otherwise unremarkable. The oldest child was born in 1986. Her audiogram showed a rising configuration. Acoustic middle ear muscle reflexes were absent. Distortion product evoked otoacoustic emissions (DPOAEs) were present at three frequencies in both ears, whereas transient evoked otoacoustic emissions (TEOAEs) were absent. Fluctuation in hearing level between tests was observed in the second child who was born in 1987. Acoustic middle ear muscle reflexes, DPOAEs and TEOAEs were absent in both ears. ABR, which was performed using negative, positive, and alternating polarity clicks at intensities of 100 dBnHL, showed no response in these two children. A flat/bowl shape audiometric configuration was seen in the youngest child who was born in 1998. DPOAEs were present in both ears. TEOAEs were present in the right ear, but absent in the left. Prolonged waves III and V were obtained using the negative and positive polarity stimuli but ABR recording with alternating polarity showed no response to 100 dBnHL. Speech perception ability was severely impaired in all siblings. The hearing of the parents was within normal limits.
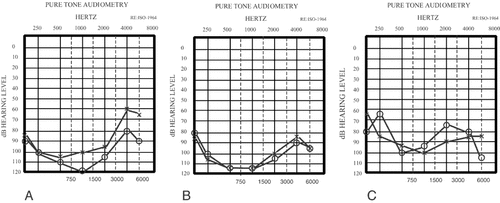
Pure tone audiograms of individuals V:1 at 8 years of age (A); V:2 at 7 years of age (B); V:3 at 6 years of age (C).
DISCUSSION
We identified a causative DNA change in only 1 out of 12 Turkish families with autosomal recessive deafness during our search for mutations in 11 known deafness loci. Earlier studies have shown that mutations in GJB2 is a major factor explaining approximately 20–25% of all nonsyndromic multiplex (mostly recessive) cases in Turkey [Baris et al., 2001; Tekin et al., 2001, 2005; Bayazit et al., 2003; Uyguner et al., 2003]. The overall rate of parental consanguinity is 25% in Turkey [Tuncbilek, 2001], but it rises up to 55% when families in which deaf probands were found to be negative for GJB2 mutations [Tekin et al., 2003]. This high rate of consanguinity suggests the presence of rare autosomal recessive alleles in the majority of Turkish families with deafness. When families are from small isolated towns of Anatolia, the genetic causes of deafness are expected to be heterogeneous. Our results, in this study, support this assumption and indicate that it is unlikely to find another common deafness gene in the Turkish population.
The c.3032T > C (p.Leu1011Pro) change in OTOF is very likely to be pathogenic because (1) it segregates with deafness in our family; (2) it was negative in 100 chromosomes of normal hearing individuals; and (3) it affects a conserved aminoacid (Table I) in a functional domain of otoferlin. Multiple isoforms of OTOF have been demonstrated, but leucine at position 1011 in the full length form is present in both short and long isoforms (www.ensembl.org). The p.Leu1011Pro mutation occurs in the fourth of the six C2 domains (C2D) in the full length of otoferlin. C2 domains are composed of two four-stranded β sheets and have highly similar structures [Yasunaga et al., 2000]. The last four C2 domains of otoferlin are predicted to bind Ca2+ [Yasunaga et al., 1999] and proteins with C2 domains are known to interact with phospholipids and proteins, with involvement in the fusion of the synaptic vesicles to the plasma membrane. Thus, otoferlin has been hypothesized to be involved in the Ca2+ triggered synaptic vesicle-plasma membrane fusion [Yasunaga et al., 2000]. It is, therefore, plausible to predict that the missense change that we found in a conserved aminoacid in the C2D domain of otoferlin impairs its function, similar to the effects of two previously reported missense mutations located in C2 domains of OTOF [Migliosi et al., 2002; Mirghomizadeh et al., 2002].
Although OTOF is a large gene with 48 coding exons, only 16 mutations causing deafness have been reported to date (Table II). Interestingly, seven of these mutations have been recently found in patients with auditory neuropathy-auditory dissynchrony (AN/AD) [Rodriguez-Ballesteros et al., 2003; Varga et al., 2003]. AN/AD is a disorder characterized by impairment of peripheral auditory function with the preservation of outer hair cell integrity. The lesion could be localized at the level of inner hair cells, the auditory nerve fibers, or the intervening synapse. Auditory neuropathy has been associated with several disorders such as neonatal hyperbilirubinemia, respiratory distress syndrome, mitochondrial disorders, peripheral neuropathies but may also be seen in the absence of any known risk factors [Cone-Wesson and Range, 2000]. The hearing loss can be congenital or late onset and the hearing level in patients with auditory neuropathy can vary from mild to profound and speech perception ability is severely impaired out of proportion to the pure tone threshold [Doyle et al., 1998]. Audiogram configurations are usually flat, but a rising audiometric configuration can be seen (28%). Hearing loss may be stable (36%) or fluctuating (29%) [Rapin and Gravel, 2003]. Otoacoustic emissions are preserved, and this indicates that outer hair cell function is normal. The incidence of auditory neuropathy within the population without risk factors is not yet established [Madden et al., 2002]. The pattern of audiological findings in two children in our family (normal otoacoustic emissions, prolonged latency of all ABR waves, impaired pure tone threshold) suggests that auditory neuropathy is the most likely diagnosis. However, OAEs were absent in the other child. Since she has been using bilateral hearing aids for several years, emissions might have disappeared as a consequence of damage from hearing aid use. Although the phenotype associated with OTOF mutations was reported as being sensorineural hearing loss before the demonstration of OTOF mutations in individuals with AN/AD in 2003, there is a good possibility that AN/AD is the only phenotypic manifestation of OTOF mutations. Diminishing OAE responses as the child gets older and insufficient evaluation of affecteds may have led to the missing of the diagnosis of AN/AD.
| Mutation | Effect on protein | Reference |
|---|---|---|
| Affecting only one aminoacid | ||
| [c.1469C > A; c.1544T > C], [p.Pro490Gln;p.Ile515Thr] | C2C domain |
Mirghomizadeh et al. [2002] |
| c.3032T > C (p.Leu1011Pro) | C2D domain | This study |
| c.5473C > G (p.Pro1825Ala) | C2F domain |
Migliosi et al. [2002] |
| c.5816G > A (p.Arg1939Gln) | Carboxy terminus |
Varga et al. [2003] |
| c.5860-5862delATC (p.Ile1954del) | Carboxy terminus |
Rodriguez-Ballesteros et al. [2003] |
| c.5960C > G (p.Pro1987Arg) | Carboxy terminus |
Varga et al. [2003] |
| Multiple affected aminoacids | ||
| c.709C > T (p.Arg237Ter) | Truncation |
Houseman et al. [2001] |
| c.711-2A > G | Exon skipping; truncation |
Yasunaga et al. [2000] |
| c.1651delG (p.Glu551fsTer4) | Truncation |
Varga et al. [2003] |
| c.2122C > T (p.Arg708Ter) | Truncation |
Rodriguez-Ballesteros et al. [2003] |
| c.2485C > T (p.Gln829Ter) | Truncation |
Migliosi et al. [2002] |
| c.2866 + 1G > A | Exon skipping or frameshift |
Adato et al. [2000] |
| c.4275G > A (p.Trp1425Ter) | Truncation |
Rodriguez-Ballesteros et al. [2003] |
| c.4362 + 2T > G | Exon skipping or frameshift |
Rodriguez-Ballesteros et al. [2003] |
| c.4491T > A (p.Tyr1497Ter) | Truncation |
Yasunaga et al. [2000] |
| c.4799 + 1G > C | Exon skipping or frameshift |
Varga et al. [2003] |
Since evaluations for AN/AD are not routinely performed on every individual with hearing loss, audiogram shapes are sometimes helpful for identifying genetic causes. Our study demonstrates that persons with OTOF mutations may have better thresholds at high frequencies on pure tone audiograms or fluctuation in hearing level between tests, as might be expected in AN/AD. It should be noted that four other individuals with OTOF mutations have been reported to have bowl shaped audiograms [Varga et al., 2003], supporting the idea that this type of otherwise uncommon audiograms can be associated with mutations in OTOF.
Acknowledgements
This study was supported by the Ankara University Scientific Research Projects (Grant# 20030809103) and Turkish Academy of Sciences in the framework of the young scientist award program (MT/TUBA-GEBIP/2001-2-19) to MT. We are grateful to Dr. Susan Kupka from the University of Tübingen, Germany for providing us oligoprimers, which were partially used in this study.





