Search for somatic 22q11.2 deletions in patients with conotruncal heart defects
Abstract
A wide range of clinical variability in patients with 22q11.2 deletions has been demonstrated in numerous studies. Nevertheless, it is still an open question if major genetic factors contribute to clinical expression. Therefore one aim of this study was to investigate, if patients with 22q11.2 deletion and conotruncal heart defects show a “second hit” somatic 22q11.2 deletion in tissue from the conotruncus, heart vessels or thymus. The second aim was to analyse patients with conotruncal heart defects without 22q11.2 deletion in blood cells for somatic deletion mosaicism. We were able to study tissue samples from heart surgery from 23 patients, 9 of whom had 22q11 deletions by FISH analysis on metaphase spreads from peripheral lymphocytes. Analysis of 18 polymorphic markers from the 22q11.2 region in DNA prepared from thymus and/or heart vessels and/or conotruncus tissue and peripheral lymphocytes in each patient did not show any allelic loss. Thus somatic 22q11.2 deletions apparently do not play a major role in conotruncal heart defects in patients with or without germ line 22q11.2 deletion. © 2003 Wiley-Liss, Inc.
INTRODUCTION
Deletion 22q11.2 is a major cause of congenital heart disease (CHD), accounting for about 5% of all CHD [Wilson et al., 1994], up to 15% of conotruncal CHD and up to 20% in foetuses with CHD. However, despite of a common deletion size of 3 Mb patients with a 22q11.2 deletion have a highly variable phenotype [Carlson et al., 1997b]. The underlying mechanism is still an open question, although imprinting, unmasking of recessive mutations by hemizygosity, unbalanced regulatory effects, a second-hit theory and environmental factors have been considered as modifying factors [Hall, 1993; Dallapiccola et al., 1996; Hatchwell, 1996]. As both, the common and the atypical deletions are mediated by several low copy repeats within the region [Edelmann et al., 1999a,b; Rauch et al., 1999; Saitta et al., 1999; Shaikh et al., 2001; Garcia-Minaur et al., 2002], our hypothesis was that an additional somatic 22q11.2 deletion might be a frequent event, modifying the phenotype in terms of a second-hit-theory. Furthermore somatic 22q11.2 deletions may explain atypical phenotypes in patients with CHD [Consevage et al., 1996]. To prove these two hypotheses, we analysed 38 probes from thymus and/or heart tissue for somatic 22q11.2 deletions from 23 patients with conotruncal CHD.
MATERIALS AND METHODS
We analysed children with congenital conotruncal heart defects undergoing heart surgery whose parents had agreed to a collection and study of tissue if available during surgery. To determine germ line 22q11.2 status, FISH analysis was performed on metaphase spreads from peripheral blood cells with 10 DNA probes covering the common and atypical deletion regions (Fig. 1): 6E8 (D22S427) [Edelmann et al., 1999a], 51H3 (D22S1649) [Carlson et al., 1997a], 70A2 (D22S1694) [Carlson et al., 1997a], Pac 140D4 (HIRA) [Carlson et al., 1997a], co23 (UFD1L) [Pizzuti et al., 1997], D0832 (COMT) [Carlson et al., 1997a], 48c12 (D22S264) [Edelmann et al., 1999b], cHKAD26 (D22S935) [Kurahashi et al., 1994; Kurahashi et al., 1997], 109G12 (109G12) [Edelmann et al., 1999a], BAC 438P22 (D22S425) [Rauch et al., 1999] (Fig. 1). Probes were directly labelled with Cy3 by nick translation and co-hybridised with a 22q subtelomeric control probe (GS-98-C4, locus 22QTEL31 [Knight et al., 2000]) directly labelled with FluoroX as described earlier [Rauch et al., 2001].
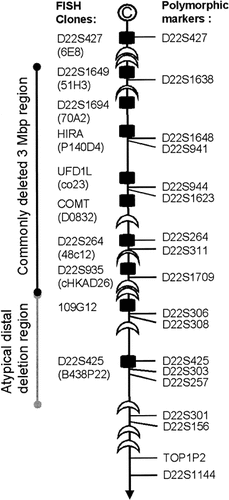
Scheme of the DiGeorge syndrome region in 22q11.2. Black rectangles represent location of probes used for FISH analysis, half moons represent low copy repeats.
To investigate somatic deletions DNA was prepared from blood lymphocytes and several tissues (heart, arteries, thymus) immediately frozen during heart surgery, and used separately for PCR amplification of 18 polymorphic markers. Amplicons of 12 and 6 markers, respectively, were pooled together using three different fluorescent dyes on a capillary sequencer as described earlier [Rauch et al., 2001].
RESULTS
We were able to study 23 children with CHD, 9 of whom with a germ line 22q11.2 deletion by FISH on peripheral blood cells (Table I, Fig. 2). As in all of these nine patients the deletion encompassed probes for loci D22S1649, D22S1694, HIRA, UFD1L, COMT, D22S264 and D22S935, but not D22S427, 109G12 and D22S425, the deletion size is in accordance with the commonly deleted 3 Mb in all of them. Results of microsatellite analysis are shown in Tables II and III and Figure 2. Allelic loss was not observed in patients with germ line 22q11.2 deletion (Table II) nor in patients without 22q11.2 deletion (Table III).
| Type of congenital heart defect (CHD) | No of patients with germ line 22q11.2 deletion | No of patients without germ line 22q11.2 deletion |
|---|---|---|
| Tetralogy of fallot (TOF) | 1 | 2 |
| Pulmonary atresia (PA-VSD) | 3 | 4 |
| Ventricular septal defect (VSD) | — | 1 |
| Interrupted aortic arch (IAA) | 3 | 3 |
| Double aortic arch (DoAA) | 1 | — |
| Truncus arteriosus communis (TAC) | 1 | 2 |
| Transposition of great arteries (TGA) | — | 1 |
| CHARGE association with ASD/VSD | — | 1 |
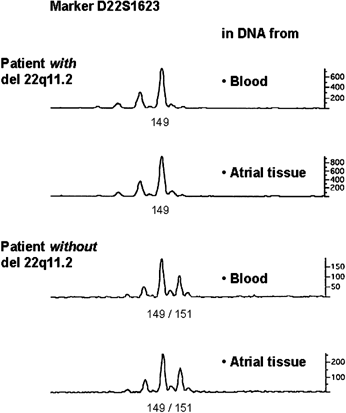
Examples of microsatellite plots.
| Marker | TOF | PA-VSD-1 | PA-VSD-2 | PA-VSD-3 | IAA type B-1 | IAA type B-2 | IAA type B-3 | DoAA | TAC type A1 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | P | B | P | B | P | B | V | B | Da | Dp | Dm | V | B | D | V | B | Da | Dp | V | B | D | B | P | |
| D22S427 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 |
| D22S1638 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D22S941 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D22S1648 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D22S944 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D22S1623 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D22S264 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D22S311 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D22S1709 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D22S306 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D22S308 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 |
| D22S425 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 |
| D22S303 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 |
| D22S257 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 |
| D22S301 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D22S156 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 |
| TOP1P2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| D22S1144 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Marker | TOF-1 | TOF-2 | PA-VSD-1 | PA-VSD-2 | PA-VSD-3 | PA-VSD-4 | VSD-1 | IAA type A-1 | IAA type A-2 | IAA type B | TAC type A2 | TAC type A4 | TGA | CHARGE | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | T | P | B | T | P | B | T | B | T | P | B | T | D | B | T | A | B | P | B | V | B | T | A | B | T | G | B | G | B | D | B | T | D | M | B | P | |
| D22S427 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 |
| D22S1638 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 |
| D22S941 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 |
| D22S1648 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D22S944 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D22S1623 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 1 | 1 |
| D22S264 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 |
| D22S311 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D22S1709 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 |
| D22S306 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D22S308 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 |
| D22S425 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 |
| D22S303 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 |
| D22S257 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| D22S301 | 2 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| D22S156 | 2 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 |
| TOP1P2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 |
| D22S1144 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 3 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
- B, DNA from blood; T, DNA from thymus; G, DNA from great artery; A, DNA from aorta; P, DNA from pulmonic art; D, DNA from Ductus Botalli (Da, arterial part; Dp, pulmonic part; Dm, middle part); V, DNA from atrial septum; M, DNA from heart muscle.
DISCUSSION
As neural crest ablation studies produced abnormalities of the type seen in DiGeorge syndrome this condition is considered a neurocristopathy [Van Mierop and Kutsche, 1986; Kirby and Waldo, 1990]. The cardial neural crest cells migrate via the 3rd, 4th and 6th pharyngeal arches to the outflow tract of the developing heart, and continue to participate in the formation of the aorticopulmonary and truncal septa [Kirby and Waldo, 1990]. The 3rd, 4th and 6th pharyngeal arches constitute parts of the aortic arch, of the carotids and right subclavian arteries, the ductus arteriosus, the thymus, the thyroid and the parathyroids [Kirby and Waldo, 1990; Larsen, 2001]. Therefore we considered a second hit deletion to be detectable if present in the latter tissues. Although diverse origins of cells within the analysed tissue would dilute second hit deletions, and low level mosaicism within the analysed tissues would not be detectable by our approach, our results show that there is no evidence for somatic 22q11.2 deletions in patients with conotruncal heart defects with or without germ line 22q11.2 deletion. Therefore we conclude that somatic 22q11.2 deletions in pharyngeal arch derived tissue do not play a major causative role in patients with unexplained conotruncal heart defects.
With reference to patients with germ line 22q11.2 deletion, our results exclude second hit deletions as a common mechanism to explain clinical variability concerning the manifestation of a CHD.
Observations of phenotypic discordance especially in heart manifestation in monozygotic twins concordant for 22q11.2 deletion has led some authors to postulate that the clinical variability in this disorder is generally not caused by genetic factors [Goodship et al., 1995; Fryer, 1996; Hatchwell, 1996; Yamagishi et al., 1998; Vincent et al., 1999; Lu et al., 2001]. As we observed monozygotic twins concordant for 22q11.2 deletion and conotruncal heart defects, we concluded that known teratogenic effects of twinning and twin pregnancy itself were likely to contribute to the discordance observed by others, and that in singletons it is most likely that genetic background contributes to variability [Rauch et al., 1998]. Our view is in line with that of Singh et al. [2002], who stress the possibility of differences of methylation patterns depending on the stage of twinning as epigenetic modification.
However, recent studies on Df1/+ mice, which model 22q11.2 deletion due to Tbx1 haploinsufficiency, showed that the penetrance of cardiovascular defects varies widely in different genetic backgrounds, thus demonstrating for the first time a major genetic control over phenotypic variability of 22q11.2 deletion [Taddei et al., 2001]. Subsequently, Vitelli et al. [2002] demonstrated that Tbx1+/−;Fgf8+/− mutants present with a significantly higher penetrance of aortic arch artery defects than Tbx1+/−;Fgf8+/+ mutants, while Tbx1+/+;Fgf8+/− animals are normal. Moreover, preliminary results of an association study between the expression of CHD in 22q11.2 deletion patients and SNPs within the promotor region of VEGF revealed a significantly increased risk for CHD associated with a certain haplotype [Stalmans et al., 2003].
Thus, it is likely that there are major genetic modifiers of penetrance of cardiovascular defects in human individuals with chromosome 22q11.2 deletions, although our study shows that somatic second hit deletions are at least not major modifiers.
Acknowledgements
We thank the families for their kind co-operation, Brigitte Schenker for skilful technical assistance, and Bernice Morrow, Peter Scambler, Giuseppe Novelli, Jonathan Flint and the Japanese Cancer Research Resources Bank for providing DNA clones.




