Previously undescribed nonsense mutation in SHH caused autosomal dominant holoprosencephaly with wide intrafamilial variability
Abstract
Holoprosencephaly (HPE) is the most common developmental defect of the forebrain and midface in humans, with a frequency of 1/16,000 live births. Different genes are implicated in the pathogenesis of HPE; these include SHH, ZIC2, SIX3, TGIF, and human DKK1. We describe here a family with recurrence of autosomal dominant HPE in different members showing a wide clinical variability. The mother presents a single central maxillary incisor and mild hypotelorism as signs of the diseases, while three of her sons were affected by HPE. By direct sequencing and restriction analysis of exon 2 of the SHH gene, we have identified a previously undescribed nonsense mutation at codon 128 (W128X). The identification of this mutation allowed us to give a prenatal diagnosis in this family and confirms a wide intrafamilial variabilty in the phenotypic spectrum. © 2003 Wiley-Liss, Inc.
INTRODUCTION
Holoprosencephaly (HPE) is the most common developmental defect of the forebrain and midface in humans [Matsunaga and Shiota, 1977]. It is a genetically and phenotypically heterogeneous disorder with a frequency of 1/16,000 live births, and it represents a frequent cause of prenatal death in humans, detected in 1/250 induced abortions [Cohen, 1989a]. The clinical and etiologic heterogeneity of HPE has been well documented [Muenke, 1989; Cohen, 1989b]. Clinical expression is variable: in the most extreme cases, anophthalmia or cyclopia is evident, along with a congenital absence of the mature nose, while in the less severe form, the features are facial dysmorphism characterized by ocular hypertelorism, defects of the upper lip and/or nose, and absence of the olfactory nerves or corpus callosum. Several intermediate phenotypes involving both the brain and face have been described [Muenke, 1994].
Several genes are implicated in the pathogenesis of HPE: 12 different loci have been associated with HPE and several distinct human genes have been identified as responsible for HPE [Roessler and Muenke, 1998]. These include sonic hedgehog (SHH) [Belloni et al., 1996; Roessler et al., 1997; Odent et al., 1999], ZIC2 [Brown et al., 1998], SIX3 [Schell et al., 1996; Wallis and Muenke, 2000], TG-interacting factor (TGIF) [Gripp et al., 2000], and human DKK1 [Roessler et al., 2000].
So far, a total of 46 mutations have been identified in 364 unrelated HPE patients, corresponding to a mutation identification rate of 12.6% [Wallis and Muenke, 2000]. In particular, 26 different mutations in SHH have been described in 27 unrelated patients, 7 different mutations in ZIC2 have been described in 11 affected individuals, and 4 different mutations have been identified in 4 unrelated patients in each SIX3 and TGIF [Wallis and Muenke, 2000]. Despite the fact that sporadic HPE is more frequent than familial, SHH mutations occur more often in familal HPE. In the paper by Wallis and Muenke [2000], SHH mutations were detected in 3.7% of sporadic cases and in 18% of familial cases. Moreover, the mutation detection rate was of 37% considering only families with autosomal transmission of HPE. The other genes analyzed by Wallis and Muenke [2000] account for 1% (SIX3 and TGIF) to 5% (ZIC2) in the pathogenesis of sporadic and familial HPE.
The role of SHH in the pathogenesis of HPE has been further clarified. The SHH gene encodes a predicted protein with 92.4% identity to its mouse homolog, expressed in fetal intestine, liver, lung, and kidney, but undetectable in adult tissues examined. Targeted disruption of the Shh gene in mice causes midline structural defects, lack of ventral forebrain development, limb anomalies, and cyclopia [Chiang et al., 1996]. The SHH gene has been mapped to chromosome 7q [Marigo et al., 1995; Belloni et al., 1996], and mutations in the cytogenetic locus 3 of HPE (HPE3) patients were first reported by Roessler et al. [1996].
Interestingly, three patients with SHH mutations also had abnormalities in other genes that are expressed during forebrain development [Nanni et al., 1999]. These data suggest that, given the great intrafamilial clinical variability in kindreds carrying an SHHmutation, other genes acting in the same or different developmental pathways might function as modifiers for expression of the HPE spectrum.
We report here a family with recurrence of autosomal dominant HPE in different members showing a wide clinical variability. The identification of a previously undescribed SHH mutation allowed us to confirm the inheritance of the disorder and to perform molecular prenatal diagnosis in the family.
MATERIALS AND METHODS
Clinical Reports
A holoprosencephalic female (Fig. 1b) was born to a 23-year-old mother with a single central maxillary incisor (Fig. 1a) and a 26-year-old apparently healthy father. The parents were not consanguineous. The newborn had microcephaly, hypotelorism, cebocephaly, palatoschisis, and micrognathia. Chromosomes were normal. Computed tomography (CT) of her brain showed alobar HPE. The mother was of normal intelligence and stature, and her brain CT scan was normal. These cases were reported by Camera et al. [1992].
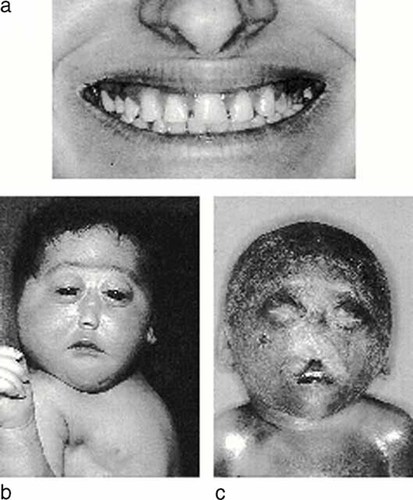
a: Single central maxillary incisor in the 23-year-old mother. b: Holoprosencephalic female showing microcephaly, hypotelorism, cebocephaly, palatoschisis, and micrognathia. c: Fetus presenting microcephaly, premaxillary agenesis with hypotelorism, flat boneless nose, and a median pseudocleft of the upper lip.
During the third pregnancy, at 21 weeks of gestation, fetal ultrasonographic examination revealed the presence of HPE. The pregnancy finished at 22 weeks with a miscarriage. The fetus had microcephaly, premaxillary agenesis with hypotelorism, flat boneless nose, and a median pseudocleft of the upper lip (Fig. 1c). Karyotype was normal (46, XY) and DNA was extracted from fetal tissues.
We identified a nonsense mutation at codon 128 (W128X) (see Results and Discussion sections) in this fetus and in the mother, by direct sequencing and restriction analysis of exon 2 of the SHH gene.
In an additional pregnancy, a chorionic villus sampling was performed at 11 weeks of gestation and sequencing analysis of the fetus DNA showed the W128X mutation. The parents were informed of our inability to make a correct phenotypic diagnosis at that period of gestation; due to the wide intrafamilial variability, we were not able to know whether the fetus had alobar HPE as his sibs or a single central maxillary incisor as his mother. The pregnancy was terminated and the autopsy showed alobar HPE.
SHH Analysis
The SHH gene was amplified as already reported [Roessler et al., 1996, 1997]. Direct sequencing was performed using dye terminator chemistry (ABI Prism, Big Dye Terminator Cycle Sequencing kit, Applied Biosystems, Foster City, CA), following the user manual instructions. The electrophoresis of the cycle-sequencing products was carried out in an ABI 377 automatic sequencer (Applied Biosystems, Foster City, CA). The genotyping of the family was carried out using polymorphic markers (D7S550, D7S2465, D7S2423) spanning the region containing the SHH locus. PCR was carried out under standard conditions. PCR products labeled with a fluorescent amidite were loaded in an automated sequencer ABI 377 (Applied Biosystems, Foster City, CA), and allele size was defined using Genescan software.
RESULTS AND DISCUSSION
In the present report we have identified a previously undescribed SHH mutation in a family with recurrence of autosomal dominant HPE. This mutation consists of a G->A transition at cDNA nucleotide 383, causing a substitution of a stop codon (TAG) for a tryptophan (TGG) at codon 128 in exon 2 (W128X) (Fig. 2a).
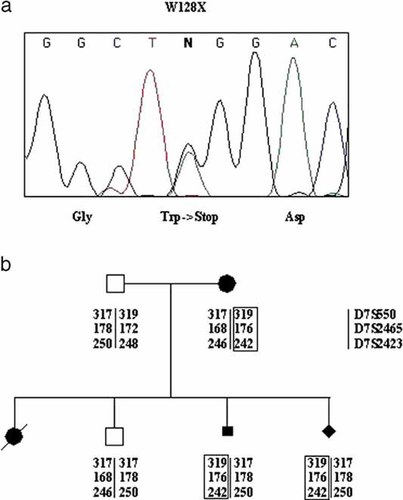
a: Sequence showing the G->A transition that causes the W128X substitution. b: Segregation of an “at-risk” haplotype in the family members.
The segregation in the family was confirmed by restriction analysis. The mutation introduces a new MaeI site in the amplification product containing exon 2, resulting in a 163-bp band in the affected individuals (data not shown). Furthermore, the genotyping of the family with polymorphic markers (D7S550, D7S2465, D7S2423) spanning the region containing the SHH locus showed the segregation of an “at-risk” haplotype (Fig. 2b).
The identification of the causative mutation in this family allowed us to confirm that the mother is a carrier with a 50% chance of transmitting this alteration to her children.
The identification of this mutation allowed prenatal diagnosis: transabdominal chorionic villus sampling was performed at 11 weeks of gestation. In order to exclude the presence of fetal DNA contamination by maternal tissues, haplotype analysis was carried out using polymorphic markers from the SHH locus. This analysis excluded maternal contamination of the DNA extracted from the chorionic villus sample and confirmed the presence of the “at-risk” haplotype in the fetus. The pregnancy was terminated, and the analysis of the fetus confirmed the presence of HPE.
Other families with recurrence of autosomal dominant HPE with variable expressivity have been already observed [Wallis and Muenke, 2000]. The phenotype of HPE is quite variable even within the same family and proceeds in a continuous spectrum from severe manifestations with major brain and face anomalies to clinically normal individuals [Cohen, 1989b; Muenke, 1994; Ming and Muenke, 1998]. Cantu et al. [1978] reported a patient showing at birth an aggregate of craniofacial malformations consisting of microbrachycephaly, ocular hypotelorism, severe frontonasal hypoplasia, and wide median cleft lip and anterior palate. The mother showed microcephaly and persistence of both decidual canines without visible secondary ones, which were detected developed within the bone and directed toward the midline, each ending above the corresponding central incisor tooth. In this case, the presence of dental abnormalities was explained as a manifestation of HPE, and this observation permitted the differentiation of an autosomal dominant form from sporadic HPE cases. Similarly, in the family described in this paper, the presence of a single central maxillary incisor tooth in the mother of affected HPE children was considered a hallmark of a dominant form. This hypothesis has been confirmed by the identification of a SHH nonsense mutation in the mother and in two affected HPE sons. The wide variability of the phenotypic spectrum in the observed affected individuals carrying the same stop codon mutation could probably be explained by the presence of interaction of multiple gene products and/or environmental factors. Furthermore, the finding of SHH mutations in only a minority of the total cohort of HPE patients underscores the significant etiologic heterogeneity of this condition. Potentially, defects in any genes involved in the developmental pattern of the forebrain could be involved in the pathogenesis of HPE, and in particular, genes interacting with the SHH pathway, such as PTC and SMO as well as the GLI genes, are considered not only good candidates but also modifiers [Wallis and Muenke, 2000].
Recently, it was described that SHH sets off a chain of events in target cells, leading to the activation and repression of target genes by transcription factors in the Gli family [Villavicencio et al., 2000]. Dysregulation of the SHH-PTCH-GLI pathway leads to several human diseases, including birth defects and cancers [Villavicencio et al., 2000]. Defects in any of several steps in the SHH-PTCH-GLI pathway lead to similar clinical phenotypes, presumably through functionally equivalent effects on downstream targets genes.




