Two-loci ADA haplotypes in autistic disorder
To the Editor:
Among neuropsychiatric disorders, autism (AD) is currently viewed as one of those with the strongest genetic component, with an underlying complex genetic susceptibility involving several loci [Bailey et al., 1995; Pickles et al., 1995].
Recently two groups [De Luca et al., 1999; Persico et al., 2000; Bottini et al., 2001] have independently reported an association, at the population level, between the common adenosine deaminase biochemical polymorphism (ADA1) and autism. The allele associated with the lower enzymatic activity, ADA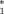 2 (G22A; Asp8Asn), was found to be significantly more frequent in three independent autistic samples when compared to their matched controls.
2 (G22A; Asp8Asn), was found to be significantly more frequent in three independent autistic samples when compared to their matched controls.
Given these results, the observed association seems unlikely to depend on chance alone or on biased controls; however, we cannot rule out that the observed results are due to a linkage disequilibrium between the ADA1 polymorphism and a causative polymorphism located in its vicinity.
To assess whether the AD-adenosine deaminase association actually encompasses the ADA1 biochemical polymorphism, we have examined three ADA polymerase chain reaction polymorphisms (PCRP) in relation to autism using pairwise measures of linkage disequilibrium. The three markers span 28Kb inside the ADA structural gene. Autistic patients and controls, as well as the ADA1 PCRP were described previously [Bottini et al., 2001]. Two more PCRPs examined here are respectively generated by the presence/absence of a PstI site (ADA2; nt19465–19470; intron 2) and of a MluNI site (ADA6; nt31230–31235; exon 6) [Hirschhorn et al., 1994]. Each locus in each sample was in equilibrium according to Hardy-Weinberg, and both markers were found to occur with the same frequency in autistic patients and in controls. Haplotype frequencies are maximum likelihood estimates (MENDEL program, Dept. of Biostatistics, University of Michigan, Ann Harbor, MI). The measure of linkage disequilibrium (i.e., the D parameter), was calculated as the product of the frequencies of the genotypes in coupling divided by the product of the frequencies of the genotypes in repulsion.
Table I reports the distribution of the two-loci haplotypes in AD samples and in controls. Only haplotypes including the ADA1 polymorphism are shown. In the population from Rome, the test provides evidence for disequilibrium between ADA2/ADA1 and ADA6/ADA1 haplotypes, both in autistic and in control samples, whereas in Sicily the disequilibrium is virtually absent in both samples. Moreover, in Rome the pattern of disequilibrium involving ADA2 and ADA1 shows differences between the autistic and control samples: in the autistic group, there is a drastic relative increase of ADA /ADA
/ADA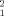 haplotype compared to controls. No such pattern was detected in the group from Sicily. Since the Sicilian population may be assumed to be relatively homogeneous, while the population of Rome is a mixture of people from various regions of Italy, we believe that the pattern of disequilibrium observed in this population represents an artifact due to stratification. This hypothesis is supported by a recent study in 51 two-generation families not showing linkage disequilibrium (Lucarelli et al., submitted for publication). Our results on two-loci haplotypes support the hypothesis that inside the ADA structural gene only the biochemical polymorphism (ADA1) confers susceptibility to autism.
haplotype compared to controls. No such pattern was detected in the group from Sicily. Since the Sicilian population may be assumed to be relatively homogeneous, while the population of Rome is a mixture of people from various regions of Italy, we believe that the pattern of disequilibrium observed in this population represents an artifact due to stratification. This hypothesis is supported by a recent study in 51 two-generation families not showing linkage disequilibrium (Lucarelli et al., submitted for publication). Our results on two-loci haplotypes support the hypothesis that inside the ADA structural gene only the biochemical polymorphism (ADA1) confers susceptibility to autism.
| Loci | AD | Controls | Difference between AD and controls F | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1/1 | 2/1 | 1/2 | 2/2 | Haplotypes | D | 1/1 | 2/1 | 1/2 | 2/2 | Haplotypes | D | ||
| Rome | |||||||||||||
| ADA2/ | 0.66 | 0.15 | 0.13 | 0.07 | 156 | 2.37 | 0.69 | 0.26 | 0.02 | 0.03 | 200 | 3.98 | 0.00003 |
| ADA1 | (0.04) | (0.03) | (0.03) | (0.02) | (0.03) | (0.03) | (0.01) | 0.02 | |||||
| ADA2/ | 0.15 | 0.66 | 0.07 | 0.12 | 156 | 0.39 | 0.21 | 0.74 | 0.02 | 0.03 | 200 | 0.43 | 0.00035 |
| ADA1 | (0.03) | (0.04) | (0.02) | (0.03) | (0.03) | (0.03) | (0.01) | (0.01) | |||||
| Sicily | |||||||||||||
| ADA6/ | 0.66 | 0.13 | 0.12 | 0.04 | 86 | 1.22 | 0.76 | 0.19 | 0.04 | 0.01 | 90 | 1.00 | NS |
| ADA1 | (0.05) | (0.04) | (0.04) | 0.03 | 0.05 | 0.04 | (0.03) | (0.02) | |||||
| ADA6/ | 0.12 | 0.71 | 0.02 | 0.15 | 86 | 1.27 | 0.19 | 0.76 | 0.01 | 0.04 | 90 | 1.00 | 0.066 |
| ADA1 | 0.04 | (0.05) | 0.02 | (0.04) | (0.04) | (0.05) | (0.02) | (0.03) | |||||
- * Only the haplotypes including the ADA1 (Asp8Asn polymorphism) are reported in the table. Standard errors are in parentheses. NS, not significant.
Bearing in mind the role of adenosine deaminase in the normal development and function of both nervous and immune systems [Franco et al., 1997; van Gent et al., 1997], the observed association of the less active ADA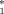 2 allele with autism justifies a specific function of the ADA gene in the pathology of autism, in conjunction with other major genes. Yet our analysis is far from being conclusive and more genetic analysis will be needed at the ADA locus, examining more markers inside and near the gene, in order to clarify the specific role of this gene and of its genetic variability in AD.
2 allele with autism justifies a specific function of the ADA gene in the pathology of autism, in conjunction with other major genes. Yet our analysis is far from being conclusive and more genetic analysis will be needed at the ADA locus, examining more markers inside and near the gene, in order to clarify the specific role of this gene and of its genetic variability in AD.




