Complex karyotype in a polycythemia vera patient with a novel SETD1B/GTF2H3 fusion gene
Conflict of interest: Nothing to report.
A physician or group of physicians considers presentation and evolution of a real clinical case, reacting to clinical information and data (boldface type). This is followed by a discussion/commentary
Abstract
The patient had been diagnosed with polycythemia vera (PV) in 1999, at the age of 61, according to the criteria of the Polycythemia Vera Study Group (PVSG) on the basis of the increased red cell mass by isotope determination, normal oxygen saturation, low plasma erythropoietin level, presence of endogenous erythroid colonies (EEC), and splenomegaly. Histopathology of bone marrow biopsy was also consistent with polycythemia vera with no evidence of increased reticulin fibrosis. A karyotype analysis was not performed at that time. He had been treated initially with phlebotomies and then with hydroxyurea with the aim to obtain a better control of hematocrit; he was under low-dose aspirin. In 2009, 10 years after the diagnosis, while the patient was still being treated with hydroxyurea and phlebotomies, he noticed worsening of general conditions and fatigue, and the appearance of night sweats; he also reported that his spleen volume had increased rapidly in the past few months. He complained of severe pruritus especially after (but not limited to) a shower. He was referred to our center for further evaluation. At presentation, his blood counts were as follows: hemoglobin 157 g/L, hematocrit 54.7%, leukocytes 13.1 × 109/L, platelets 238 × 109/L, LDH 856 U/L (normal upper limit, 250 U/L). Blood film examination showed neutrophilia (8.9 × 109/L) but immature myeloid cells and nucleated erythroblasts were absent. The spleen was 14 cm below the left costal margin, the liver was at 4 cm below the right costal margin. He was found to harbor the JAK2V617F mutation with an allele burden of 85% and the circulating CD34+ cell count was 14 × 106/L. A bone marrow biopsy showed the presence of hyperplasia of myeloid and erythroid lineages, increased number of scattered megakarocytes without overt morphologic abnormalities; reticulin fibrosis was grade 1 according to the European classification. On these basis, we considered the patient as presenting the features of PV according to the 2008 WHO classification of myeloid neoplasms associated with grade 1 reticulin fibrosis. Am. J. Hematol. 89:438–442, 2014. © 2014 Wiley Periodicals, Inc.
The patient was initially diagnosed with polycythemia vera (PV) according to the Polycythemia Vera Study Group (PVSG) criteria 1, 2, and this diagnosis was reconfirmed 10 years later using the 2008 updated WHO criteria, that included the JAK2V617F mutational status 3. His clinical course had been substantially uneventful without major cardiovascular events, no hemorrhages, but characterized by long-lasting and disabling pruritus and the difficulty to control hematocrit level with phlebotomy alone, that prompted the introduction of hydroxyurea in the therapeutic armamentarium. After several years, clinical signs suggesting progression of the disease were noticed, but the results of hematologic and histopathological re-evaluation supported again the diagnosis of PV. An increase in reticulin fibrosis, with a loose network of reticulin with intersections, especially in perivascular areas 4, was noticed in the bone marrow, compatible with grade 1 of 3 (European scale). At diagnosis, about 14% of the patients with PV display grade 1 reticulin fibrosis, associated with palpable splenomegaly and correlated negatively with the occurrence of thrombotic events during the disease course 5. It has been reported that initial bone marrow reticulin fibrosis increased of 2.5-fold the risk of developing postpolycythemia vera myelofibrosis (PPV-MF) 5; of note, in this particular patient, reticulin fibrosis was first reported after 10 years from the original bone marrow biopsy. An attempt to increase the dose of hydroxyurea to 2.0 g/d in the last months had resulted ineffective because the patient developed thrombocytopenia (50 × 109/L) on several occasions, leading to hydroxyurea discontinuation. Therefore, the subject was considered as refractory/resistant to hydroxyurea according to the European Leukemia Net (ELN) criteria 6. The patient was then enrolled in a clinical trial with the histone deacethylase inhibitor givinostat, obtaining prompt relief from pruritus and rapid reduction of phlebotomies that completely stopped after 2 months 7. However, the treatment was interrupted after 6 months because of progression of a preexisting mild renal insufficiency, and the subject was shifted again to low-dose hydroxyurea (500 mg/d) plus phlebotomies.
During the diagnostic procedure, the patient also underwent cytogenetic analysis. The conventional bone marrow karyotype showed the presence of a somatic t(6;15) translocation that was absent in peripheral blood lymphocytes and was therefore considered as true somatic. An extended mutational analysis did not show additional mutations in EZH2, ASXL1, SRSF2, and IDH1/2 genes.
In addition to JAK2 or MPL mutations, that contribute to define the MPN phenotype in the large majority of patients with PV and about 60% of ET and PMF, several additional mutations have been found in genes involved in epigenetic regulation (such as ASXL1, EZH2, IDH1/2, and DNMT3A) or the spliceosome (such as SF3B1 and SRSF2) 8. Mutations in ASXL1, EZH2, SRSF2, and IDH1/2 have been reported to contribute to a negative prognostic outcome in PMF 9, but information are not yet available in patients with myelofibrosis secondary to PV or ET nor in the chronic phase of the two disorders. As a matter of fact, our patient did not present any of these prognostically adverse mutations. On the other hand, studies of MPN using classical metaphase cytogenetics have reported that less than 30% of patients exhibit an aberrant karyotype 10-12, and in most cases, these were patients with myelofibrosis. The prognostic relevance of cytogenetic aberrations acquired by patients with primary myelofibrosis warranted their incorporation in the Dynamic International Prognostic Scoring System- (DIPSS-) plus 13. Furthermore, a recent study conducted in 1,545 patients with WHO-defined PV showed a profound impact of abnormal karyotype for both overall survival and leukemia-free survival 14. Although the relatively small size of PV patients with available cytogenetic information precluded the identification of differential prognostic impact associated with specific cytogenetic abnormalities, this study clearly signified the value of obtaining cytogenetic information in patients with PV. A prognostically negative impact of karyotype aberrations other than 20q- and 13q- has been demonstrated also in patients with PPV-MF and shown to supersed the performance of clinical scores 15. More recently, by using high-resolution SNP array, it was found that JAK2V617F-positive patients have an increased number of chromosomal aberrations, particularly in secondary myelofibrosis or accelerated phase of MPN, as compared to the chronic phase 16. We were intrigued by the karyotypic aberration found in our subject, that has not been reported previously, and we aimed to characterize it further at the molecular level.
The karyotype of the patient was originally defined as: 46,XY,del(15)(q?).ish ins(6;15)(p?q2?)(PML+,wcp15+) 21. Multiple interstitial chromosome losses (involving chromosomes 6, 12, 15, and 18, respectively) were detected by SNP array CGH analysis: all were further confirmed by fluorescence in situ hybridization (FISH) with properly selected BAC probes, performed as previously described 17 (the list of the 22 FISH probes used is available upon request). To characterize further these observed aberrations, we also used High-Resolution Mate-Pair Sequencing 18.
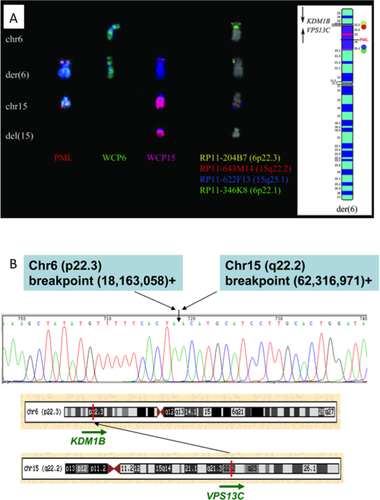
A ins(6;15) insertion, identified by FISH with a commercial probe for the PML gene (15q24) and Whole Chromosome Painting (WCP) probes of chromosomes 15 and 6 (Fig. 1A) revealed chromosome losses at all the breakpoint regions [6p22.1p22.3 (11,6 Mb), 15q21.3q22.2 (6 Mb), and 15q25.1q25.2 (3 Mb)] confirmed by SNP array CGH. Additional FISH analyses revealed that the 15q22.2q25.1 segment was inserted with an inverted orientation at 6p22 (Fig. 1A). High-resolution mate pair sequencing and genomic PCR were used to map the distal insertion breakpoint at the nucleotide level. Chromosome 6 at nt 18,163,058 was fused to chromosome 15 at nt 62,316,971, juxtaposing KDM1B (lysine (K)-specific demethylase 1B; 6p22.3) exon 4 to VPS13C (vacuolar protein sorting 13 homolog C; 15q22.2) exon 7; however, no fusion transcript resulted (Fig. 1B). The proximal insertion breakpoint joined intergenic regions (data not shown).
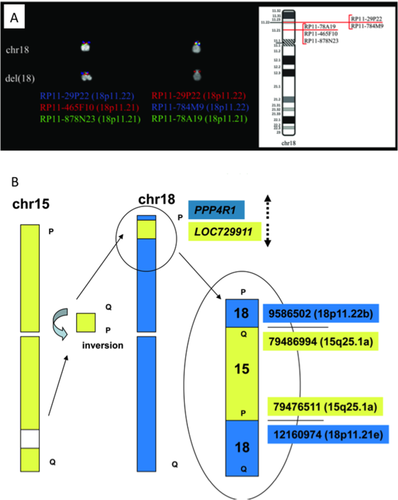
In addition, the massive sequencing approach revealed a cryptic and inverted insertion of a short chromosome 15q25.1 fragment (chr15:79,476,511–79,486,994) into chromosome 18, accompanied by a 25.7 Kb heterozygous deletion (Fig. 2A). Both fusion junctions were validated by Sanger sequencing. The distal breakpoint fused PPP4R1 (protein phosphatase 4, regulatory subunit 1; 18p11.22) to LOC7299111 (15q25.1), with opposite transcriptional orientation (Fig. 2B). Conversely, the proximal breakpoint did not involve any known gene. Furthermore, no abnormality was discovered, by using Sanger sequencing and/or Taqman gene expression assays respectively, in the genomic sequence and expression levels of the genes eventually involved in, or near, the breakpoint region or located in the deleted regions with a putative pathogenetic interest (namely, KDM1B, RFX7, NEDD4, FOXB1, and GNAL) (not shown in detail).
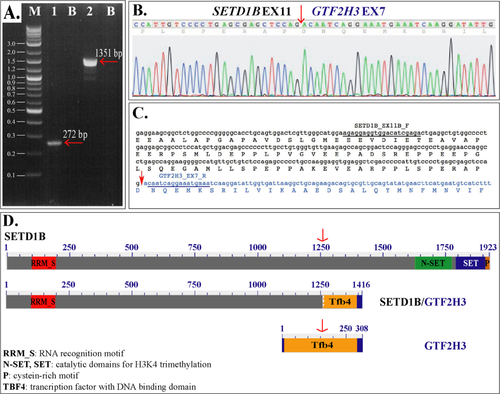
Interestingly, we identified a heterozygously deleted region at position 12q24.31, which was shown to juxtapose the SETD1B (SET domain containing 1B) gene at intron 11 (chr12:122,257,801) with the GTF2H3 (general transcription factor IIH, polypeptide 3) gene at intron 5 (chr12:124,137,254). This alteration results in both genes having the same transcriptional orientation (Fig. 3). The presence of a 5′SETD1B/3′GTF2H3 fusion gene was validated by Sanger sequencing at both DNA and RNA level. RT-PCR with combinations of SETD1B forward and GTF2H3 reverse primers successfully amplified cDNA fragments. The SETD1B_EX11B_F/GTF2H3_EX7_R and SETD1B_EX7_F/GTF2H3_EX13_R primer pairs (see legend to Fig. 3) yielded single bands of 272 and 1351 bp, respectively (Fig. 3A). The sequencing of both RT-PCR products confirmed the occurrence of a 5′SETD1B/3′GTF2H3 fusion transcript (Fig. 3B). In detail, SETD1B (accession no. NM_015048) at exon 11 was fused in frame with GTF2H3 (accession no. NM_001516) at exon 7 (Fig. 3C). In silico translation of the fusion transcript, obtained by ORF finder, showed a putative protein maintaining the SETD1B RNA binding domain (RRM_S) at its N-terminus and a portion of GTF2H3 DNA binding domain (Tfb4) at its C-terminus (Fig. 3D). It was noteworthy that mutations in SETBP1 (SET domain binding protein 1) have been recently demonstrated in about one third of atypical chronic myeloid leukemia patients and, more infrequently, in other chronic and acute myeloid neoplasms and myelodysplastic syndromes 19-21.
Because of the novelty of a gene fusion being identified in PV, we screened an additional 20 PV cases by RT-PCR for the recurrence of the 5′SETD1B/3′GTF2H3 chimeric transcript; however, no additional aberration was identified. Similarly, negative results were obtained in 44 different MPN cases (15 of PMF, 20 of ET, 5 of PPV-MF, and 4 of PET-MF).
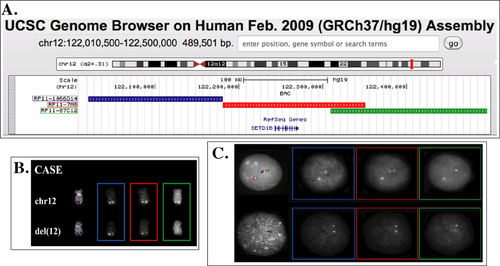
Moreover, because SETD1B is a paralog of MLL (www.ensembl.org), known for its involvement in multiple fusion genes with several partners 22, we searched for a possible recruitment of SETDB1 in additional rearrangements in PV. We properly selected, according to the latest release (February 2009: GRCh37/hg19) of the University of California Santa Cruz (UCSC) Human Genome Browser (http://genome-euro.ucsc.edu/cgi-bin/hgGateway?db=hg19), three Bacterial Artificial Chromosome (BAC) probes encompassing the whole SETD1B region (Fig. 4A). This probe set was first validated by FISH on the case under study, showing deletion of the 3′ portion of the gene on one of the two chromosomes 12 (Fig. 4B). The same FISH assay performed in 8 PV and 5 PPV-MF cases showed a splitting signal in a low percentage (3/412, 0.7%) of nuclei in only one case (Fig. 4C). This finding is in favor of a possible, although rare, rearrangement of SETD1B with other partners in PV.
Two years later, the patient showed mild anemia (hemoglobin 123 g/L) with normal ferritin levels, after having stopped phlebotomies since the last one and half year; leukocytosis (15.9 × 109/L) was increased, immature myeloid cells and erythroblasts were found in blood smears, blasts were absent. The spleen had progressed to 22 cm from the left costal margin; the patient reported loss of ≅10% of his body weight in the last months and drenching night sweats had appeared; aquagenic pruritus was still present. A bone marrow biopsy documented an overt progression to fibrosis grade 2 with reduced overall cellularity and depressed erythroid lineage, while abnormal megakaryocytes predominated and were sometimes organized in tight clusters. The patient was therefore diagnosed as post-polycythemia vera myelofibrosis (PPV-MF) according to the diagnostic criteria of the International Working Group for Myeloproliferative Disorders Research and Treatment (IWG-MRT) 23. The previously detected cytogenetic alteration was still present as the sole abnormality. The JAK2V617F allele burden was 92% and the circulating CD34+ cell count was 78.4 × 106/L.
The criteria currently used for PPV-MF diagnosis in a patients with a prior diagnosis of PV, made according to the WHO 2008 criteria, enlist the following: fibrosis grade 2–3 (on the 0–3 scale) or 3–4 (on the 0–4 scale) as obligatory criterion plus any two of: i) anemia or sustained loss of requirement of cytoreductive treatment or phlebotomy; ii) leukoerythoblastosis; iii) increase of at least 5 cm of a preexisting splenomegaly, or appearance of new splenomegaly; iv) development of at least one constitutional symptoms. The patient satisfied all above criteria, and, therefore, he was diagnosed as having PPV-MF. Because of progression of the renal insufficiency up to a chronic dialytic stage, we decided to maintain the patient on low-dose hydroxyurea. The possibility to use JAK2 inhibitors is being discussed.
In summary, we identified a novel karyotypic abnormality in a single PV patient that generated a fusion gene involving two genes, SETD1B and GTF2H3. We are aware of the limitations of the present study. First, the lack of cytogenetic information at the time of the diagnosis of PV does not allow to conclude if this rearrangement might have eventually impacted on the transformation of PV to a myelofibrotic stage. However, since ten years elapsed from the original diagnosis of PV, and considering that the transformation to PPV-MF occurred two years after the identification of the gene rearrangement, we would argue in favor of the late acquisition of such molecular abnormality speculating about a putative role in the disease evolution. Moreover, although we were unable to detected this fusion gene in an additional group of patients, a further screening for SETD1B alterations in a larger number of cases might help to definitely establish whether this gene may be recurrently involved, even at low frequency, in PV. Indeed, a recent report in a small cohort of patients with myelofibrosis, with either normal or abnormal karyotype 18, has uncovered low-frequency novel submicroscopic deletions/translocations/fusions. Finally, functional correlates of the mutant gene have not been explored in this study. However, previously mentioned findings of involvement of SETBP1 in hematologic malignancies and SETD1B in other cancers lend support to hypothesize that abnormalities of proteins containing SET domain may be associated in some respect with a myeloproliferative phenotype. Overall, these accumulating evidences underline the genetic heterogeneity of MPN, suggesting the need to unveil novel rearrangement events in addition to the most common and better known point mutations.




