Recurring homozygous ACTN2 variant (p.Arg506Gly) causes a recessive myopathy
Abstract
Objective
ACTN2, encoding alpha-actinin-2, is essential for cardiac and skeletal muscle sarcomeric function. ACTN2 variants are a known cause of cardiomyopathy without skeletal muscle involvement. Recently, specific dominant monoallelic variants were reported as a rare cause of core myopathy of variable clinical onset, although the pathomechanism remains to be elucidated. The possibility of a recessively inherited ACTN2-myopathy has also been proposed in a single series.
Methods
We provide clinical, imaging, and histological characterization of a series of patients with a novel biallelic ACTN2 variant.
Results
We report seven patients from five families with a recurring biallelic variant in ACTN2: c.1516A>G (p.Arg506Gly), all manifesting with a consistent phenotype of asymmetric, progressive, proximal, and distal lower extremity predominant muscle weakness. None of the patients have cardiomyopathy or respiratory insufficiency. Notably, all patients report Palestinian ethnicity, suggesting a possible founder ACTN2 variant, which was confirmed through haplotype analysis in two families. Muscle biopsies reveal an underlying myopathic process with disruption of the intermyofibrillar architecture, Type I fiber predominance and atrophy. MRI of the lower extremities demonstrate a distinct pattern of asymmetric muscle involvement with selective involvement of the hamstrings and adductors in the thigh, and anterior tibial group and soleus in the lower leg. Using an in vitro splicing assay, we show that c.1516A>G ACTN2 does not impair normal splicing.
Interpretation
This series further establishes ACTN2 as a muscle disease gene, now also including variants with a recessive inheritance mode, and expands the clinical spectrum of actinopathies to adult-onset progressive muscle disease.
Introduction
Alpha-actinin-2 (encoded by ACTN2) is a sarcomeric protein that is predominantly expressed in cardiac and skeletal muscle. It is an important component of the Z-disc, and it is essential for sarcomere contractility and structural stability. Alpha actinin-2 is a dimeric protein, cross-linking actin filaments with titin, and serves as a mechanosensor and scaffold for many other cytoskeletal and sarcomere-associated proteins.1, 2 Pathogenic variants in ACTN2 are a known cause of cardiac disease, including hypertrophic and dilated cardiomyopathies and arrhythmias, without reported skeletal muscle involvement.3, 4 Recently, dominantly acting monoallelic pathogenic ACTN2 variants were identified in patients with progressive skeletal myopathy with variable onset and progression without cardiomyopathy, characterized by predominant distal weakness, facial weakness in some, and histological and ultrastructural findings of a core myopathy.5-8 Recessive ACTN2 variants were implicated in skeletal core myopathy in a single case series reporting a recurring biallelic (p.Asn480Ser) ACTN2 variant in three families with adult-onset weakness.9 The underlying disease mechanism, and how monoallelic or biallelic variants in ACTN2 result in skeletal muscle and cardiac disease, remain poorly understood.
Here we report and characterize a series of patients with a novel, recurring homozygous variant in ACTN2: c.1516A>G (p.Arg506Gly), manifesting with asymmetric, progressive lower extremity predominant muscle weakness with histological findings of myofibrillar disarray and a distinct pattern of asymmetric muscle involvement on muscle MRI imaging. Taken together, we establish recessive ACTN2 variants as a cause for adult-onset skeletal myopathy, thus expanding both the phenotypic and genetic spectrum associated with this emerging group of actininopathies to now include homozygous c.1516A>G (p.Arg506Gly) ACTN2 variants.
Methods
Recruitment and sample collection
Patients were identified through their local neurology and/or genetics clinic. Informed consent for study procedures and photographs was obtained by a qualified investigator (protocol 12-N-0095 approved by the National Institute of Neurological Disorders and Stroke, National Institutes of Health; protocol 2016P001422 approved by the Mass General Brigham IRB; protocol 15-HG-0130 approved by the National Human Genome Research Institute, National Institutes of Health and protocol 11-04E approved by the CHEO Research Institute). Medical history was obtained; clinical evaluation and biopsies were performed as part of the standard diagnostic evaluation. DNA and muscle biopsy samples were obtained according to standard of care procedures. Muscle MRI was performed using conventional T1-weighted spin echo and short tau inversion recovery (STIR) of the lower extremities on different scanners at two different centers.
Genetic testing
The details of clinical and research genetic testing performed in each are listed in Supplementary 1. Confirmation of variants and segregation testing was performed by Sanger sequencing.
Haplotype analysis
To determine if shared haplotypes were present, we examined whether high-quality variants (passed Variant Quality Score Recalibration (VQSR) filtration, genotype quality >40, allele balance >0.25) within a 2.7 Mb ROH region identified in F2 (GRCh38 1:235048910-237783609) that contained ACTN2 with allele frequency <10% in gnomAD, were shared between F1 and F2.
Exontrap analysis
Cell-based splicing analysis of the mutant ACTN2 c.1516A>G was performed using Exontrap cloning vector, pET01 (MoBiTec GmbH). Template genomic DNA from patient F1P1 and a wild-type (WT) control was used to PCR amplify WT and mutant ACTN2 exon 14 and partial surrounding introns sequences (753 bp) using specific primers that included XhoI and BamHI sites (5′-CTACTCGAGGCACACAACTTCTCTGCACAT-3′ and 5′-GATGGATCCAACGGGTTTGCAACACAGAGTA-3′). PCR products were digested with XhoI and BamHI-HF restriction enzymes (New England Biolabs) and ligated to pET01 using T4 DNA ligase (New England Biolabs). Two microliter of ligation was used for transformation in TOP 10 competent cells (ThermoFisher Scientific) and selected colonies were verified by Sanger sequencing (Quintara Biosciences). HEK293T cells were cultured in DMEM (Gibco/ThermoFisher Scientific) supplemented with 10% pinnacle fetal bovine serum (FBS; Phoenix Scientific) and 1% penicillin/streptomycin (Gibco/ThermoFisher Scientific) at 37°C with 5% CO2. pET01-ACTN2-WT and pET01-ACTN2-MUT plasmids were transfected into HEK293T cells using Lipofectamine 3000 according to the manufacturer's protocol (ThermoFisher Scientific) and incubated for 24 h before RNA isolation. Total RNA was isolated from PBS-washed adherent cultured cells using RNeasy Mini kit according to the manufacturer's protocol (Qiagen). RNA (1 μg) was treated with DNAse I (ThermoFisher Scientific) and cDNA was synthesized with SuperScript IV Reverse Transcriptase (ThermoFisher Scientific) using the vector-specific primer 5′-ACTGATCCACGATGC-3′. cDNA was then PCR amplified using KAPA HiFi HotStart polymerase (Roche) and specific primers (5′-GGATTCTTCTACACACCC-3′ and 5′-CCGGGCCACCTCCAGTGCC-3′). PCR products were run on a 2.5% agarose gel and imaged using the ChemiDoc XRS+ Imaging System (Bio-Rad) and verified by Sanger sequencing.
Results
Clinical presentation
Detailed clinical information for all patients, which includes four males and three females, is summarized in Table 1, with ages ranging from 34 to 65 years at the time of the most recent examination. The muscle disease occurred sporadically in three families, only affecting a single family member. In one family (F2), there were four affected and four unaffected siblings with unaffected parents. Clinical data were not available for the deceased affected sibling in F2. All families report Palestinian ancestry. Three families (F1, F3, and F4) reported possible distant consanguinity. Family history was otherwise noncontributory (Fig. 1A).
| Family/Patient | F1P1 | F2P2 | F2P3 | F2P4 | F3P5 | F4P6 | F5P7 |
|---|---|---|---|---|---|---|---|
| ACTN2 variant | Homozygous c.1516A>G (p.R506G) | Homozygous c.1516A>G (p.R506G) | Homozygous c.1516A>G (p.R506G) | Homozygous c.1516A>G (p.R506G) | Homozygous c.1516A>G (p.R506G) | ||
| Ethnicity | Palestinian | Palestinian | Palestinian | Palestinian | Palestinian | ||
| Consanguinity | Yes | Denied | Yes | Yes | Denied | ||
| Sex | Male | Female | Female | Male | Male | Female | Male |
| Age at last examination | 44 years | 53 years | 65 years | 62 years | 61 years | 39 years | 34 years |
| First symptoms (age) | Proximal weakness (18 years) | Proximal weakness (during 20s) | Pelvic girdle weakness (teens) | Proximal LE weakness (15 years) | Proximal weakness (during 30s) | Difficulty walking (4 years) | Proximal weakness (18 years) |
| Distribution of weakness | Progressive proximal LE > UE | Progressive proximal LE > UE | Progressive proximal LE and UE weakness | Progressive proximal and distal UE and LE weakness | Progressive asymmetric proximal LE > UE; hamstrings wasting | Progressive proximal LE | Progressive asymmetric proximal and distal LE weakness |
| CK (IU/L) [reference range] | 700–1500 [52–336] | 178 [29–168] | – | – | 160–3000 | 579 [21–215] | 441–1142 |
| EMG (age) | Myopathic (31 years) | Myopathic | Myopathic, ulnar pressure neuropathy (60 years) | – | Myopathic | Myopathic (32 years) | Myopathic, primarily in proximal muscles, LE > UE (30 years) |
| Muscle biopsied (age) | R vastus lateralis (22 years) | N/A | L quadriceps (41 years) | R quadriceps (32 years) | - (39 years) | L thigh (32 years) | R quadriceps (19 years) |
| Histologic findings | Fatty replacement, fiber size variability. Lobulated and ring fibers, red rimmed vacuoles |
N/A | Myofiber atrophy, fiber size variability, Type I atrophy. Lobulated and ring fibers |
Dystrophic | Fatty replacement, fiber size variability, rimmed vacuoles | Internalized nuclei, type 1 atrophy, type 2 predominance, rimmed vacuoles | Atrophic type I and type II muscle fibers, rimmed autophagic vacuoles, suggestive of a myopathic process |
| Ultrastructural findings (on EM) | N/A | N/A | N/A | N/A | N/A | Focal aggregates of myelin figures | Sarcoplasmic vacuoles with irregular osmophilic material and membranous bodies |
| FVC % predicted (age) | 88% (44 years) | 83% (53 years) | – | – | 97% (52 years) | 83% (39 years) | 89% (33 years) |
| Cardiac evaluations (age) | Normal echocardiogram (44 years) | Normal echocardiogram (44 years) SVT during pregnancy with ablation (32 years) |
– | Second degree AVB type I, asymptomatic (58 years) | Normal echocardiogram (44 years) Normal cardiac PET scan with normal EF > 60% (55 years) |
Normal echocardiogram (36 years) Normal 13-day cardiac rhythm monitor (36 years) |
|
| Other | N/A | High arched palate | N/A | N/A | Mild ptosis (L) | High arched palate, diffuse myalgias, osteopenia | Scapular winging, sleep apnea |
- −, unknown; CK, creatine kinase; ECG, electrocardiogram; EF, ejection fraction; EM, electron microscopy; EMG, electromyography; F, female; FVC, forced vital capacity; GT, Gömöri trichrome; L, left; M, male; N/A, not available; NADH, nicotinamide adenine dinucleotide; PET, positron emission tomography; R, right; SVT, supraventricular tachycardia.
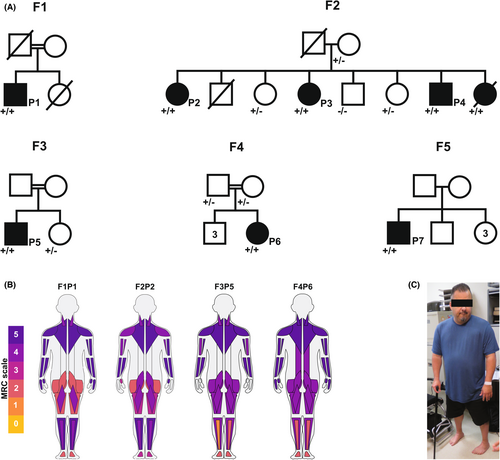
All patients presented with asymmetric, progressive, proximal and distal lower extremity weakness and to a lesser degree affecting the upper extremities. The pattern of weakness is predominantly proximal; however, a few clinical findings also pointed to distal muscle involvement including foot and/or toe drop bilaterally (Fig. 1B,C). Interestingly, two of the patients (F1P1and F2P2) were found to have finger flexor weakness as well as finger extensor weakness in digits I, II, and III more than IV and V, the reverse pattern of what is seen in MYH7-related Laing distal myopathy. Over time during adulthood, weakness was progressive with persistent loss of muscle strength and function, at times exacerbated by periods of immobility or pregnancy as reported by F2P2 and F4P6 respectively. There was no facial weakness. F3P5 had mild left-sided ptosis. Patients F2P2 and F4P6 had a high arched palate, possibly suggesting a prenatal or congenital onset of disease.
Electromyography was performed in six patients and demonstrated findings consistent with a myopathic process, with early recruitment of short duration and small amplitude motor unit action potentials. None of the patients have clinical evidence of cardiomyopathy or respiratory insufficiency at their most recent evaluation. A full description of the patients is provided in the Supplementary Materials 2.
Muscle imaging findings
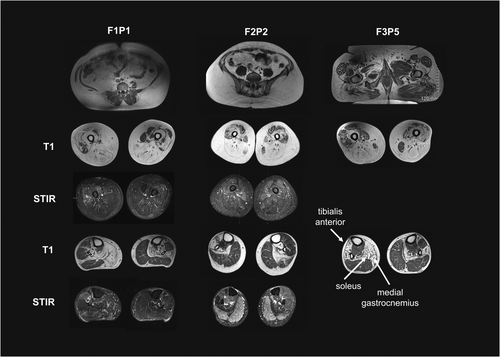
Lower extremity muscle MR imaging was available for three patients (F1P1, F2P2, and F3P5) and showed increased T1 signal consistent with fibro-fatty infiltration of muscles, reflecting an underlying myopathic disease process. In all three, there was a distinct pattern of asymmetric muscle involvement with marked fatty replacement of the posterior, medial, and gluteal compartments in the thighs (Fig. 2A). In the lower legs, there was selective involvement of the anterior tibial group and soleus with relative sparing of the gastrocnemius. A notable feature was the marked asymmetry and side-to-side variability of T1 signal change in the tibialis anterior and quadriceps muscles. Several areas of STIR hyperintensity were noted, specifically in areas of muscle with normal or near normal T1 signal (Fig. 2B), presumably highlighting areas of disease activity.
Muscle histological and ultrastructural findings
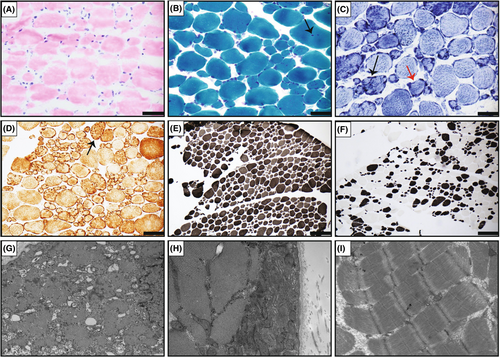
Muscle biopsies were performed in five patients (F1P1, F2P3, F3P5, F4P6, and F5P7) and demonstrated findings consistent with a chronic myopathic process, including variation in fiber size (Fig. 3A) and internalized nuclei (Fig. 3B). Lobulated and ring fibers were noted in three on nicotinamide adenine dinucleotide (NADH) and cytochrome oxidase (COX) staining (Fig. 3C, D). Overall, Type 1 fiber predominance and atrophy were noted (Fig. 3E,F). Four biopsies were noted to have rimmed vacuoles on Gömöri trichrome (GT) staining. Electron microscopy (EM) was available for two patient (F4P6 and F5P7) and revealed subsarcolemmal accumulations of mitochondria in F4P6 (Fig. 3G,H). Sarcoplasmic vacuoles with irregular osmophilic material and membranous bodies were noted in F5P7. Occasional mitochondrial aggregates were seen in association with rimmed or autophagic vacuoles. There was no evidence of disarray of Z-discs, or regional loss of mitochondria as would be seen with cores or core-like regions (Fig. 3I).
Identification of ACTN2 variants
In all families, extensive next-generation-based sequencing had been unrevealing. Exome or genome sequencing revealed one apparent homozygous missense variant in ACTN2: c.1516A>G (p.Arg506Gly) (NM_001103.2) in eight affected individuals of the five families. The variant is absent from gnomAD v2 and v3, has inconsistent in silico computational predictions (MutationTaster: disease causing; CADD: 23. PrimateAI 0.76, REVEL 0.24, AlphaMissense: 5.99E-01) and has not yet been reported to cause disease (Fig. 4A). Segregation testing was consistent with biallelic recessive inheritance in Family 2, with the variant identified in an apparent homozygous state in three affected siblings, heterozygous in two unaffected siblings and an unaffected parent, and absent in a third unaffected sibling. Parental DNA was not available in F1P1 and F3P5 for segregation testing.
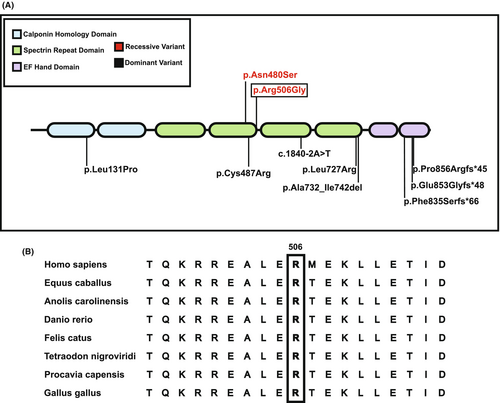
Families 1 and 2 were assessed for copy number and structural variants respectively with none identified for the ACTN2 gene. The variant (p.Arg506Gly) in ACTN2 affects a highly conserved residue (Fig. 4B). ACTN2 is constrained for loss of function (LoF) variation, with only 6 of 49.6 expected predicted LoF variants observed in gnomAD v2.1.1 (pLI score 1, LOEUF score 0.24) with 84% (CI 78–91%) of missense variation present (z-score 1.29).10 This lower-than-expected frequency of loss of function variants, indicates strong intolerance of ACTN2 to loss of function variation.
Haplotype analysis
We assessed whether Family 1 and Family 2 shared a haplotype. The (p.Arg506Gly) variant was within a region of homozygosity of 1.1 Mb in Family 1 (exome data) and 2.7 Mb in Family 2 (genome data). All homozygous variants (allele frequency <10%) from exome data in Family 1 were shared with Family 2, indicative of a shared haplotype between these two families. In addition to the causal variant, this included three variants with allele frequencies of 0.2%, 8.7%, and 3.4% in gnomAD v3.
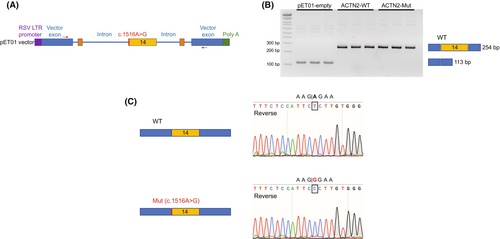
In vitro splicing analysis
The c.1516A>G variant impacts the first base pair of exon 14 of ACTN2 and thus has the potential to cause a loss of the acceptor site.11 Unfortunately, there was no muscle tissue available for RNA sequencing to evaluate whether the c.1516A>G ACTN2 variant impacts normal splicing and normal ACTN2 levels. Therefore, we pursued a cell-based splicing analysis of the mutant ACTN2 c.1516A>G using an Exontrap cloning vector (Fig. 5A). No aberrant spliced product was identified, and the mutant construct produced bands that were comparable to wild type (254 bp), indicating a normal splicing pattern (Fig. 5B). Sequence analysis of the mutant band confirmed normal splicing without deletion of exon 14 (Fig. 5C). This observation is in line with the in silico prediction provided by SpliceAI: 0.0.
Discussion
In this series, we provide detailed clinical, imaging, and histological data of seven patients from five families. Patients all manifested with a remarkably consistent phenotype of adult-onset progressive proximal, distal, and lower greater than upper extremity asymmetric weakness without significant cardiac or respiratory involvement. All patients were found to be homozygous for the identical c.1516A>G (p.Arg506Gly) ACTN2 variant. Segregation testing was pursued in two families (F2 and F4) where at least one relative was available for testing, and was consistent with a recessive mode of inheritance. This variant is rare, impacts a conserved residue, and is predicted to be damaging. All families reported Palestinian background suggesting a common origin for this variant. Haplotype testing from two families identified a shared haplotype, and thus indeed indicating a likely founder variant. Our series confirms the findings by Inoue et al (2021) who suggested a recessive ACTN2 mechanism due to a recurring c.1439A > G (p.Asn480Ser) ACTN2 variant in three families with adult-onset core myopathy. Taken together, this series expands the clinical phenotypic and genetic spectrum associated with ACTN2-related disease to now include biallelic (p.Arg506Gly) ACTN2 recessive variants as a cause for adult-onset myopathy.
Clinically, the patients in this series all present with slowly progressive and chronic muscle weakness, as is typically seen in hereditary myopathies, but also in some autoimmune myopathies such as autoimmune anti-HMGCR (3-hydroxy-3-methylglutaryl-coenzyme A reductase) myopathy.12, 13 In addition, patients were found to have predominant distal muscle weakness and marked asymmetric muscle involvement. Clinically patients with biallelic (p.Asn480Ser) ACTN2 also present with distal predominant muscle weakness, though symptom onset seems to be later in life compared to (p.Arg506Gly) ACTN2 patients, ranging from age 40–60 year compared to age 4–30 years respectively.9 Facioscapulohumeral muscular dystrophy (FSHD) commonly presents with asymmetric muscle involvement and can affect proximal and distal muscles, including anterior tibial group. However, it is most commonly an autosomal dominant (FSHD1) or digenic co-dominant (FSHD2) disease, and not consistent with the observed recessive inheritance in our patients. In addition, lack of weakness in scapular fixators and facial weakness are common features in FSHD that were absent in our patients. The proximal muscle weakness and atrophy in the lower extremities, and asymmetric tibialis anterior weakness combined with selective weakness of the wrist and finger flexors is a pattern that is reminiscence of patients with sporadic inclusion body myositis (sIBM).14 Accordingly, three patients (F1P1, F4P6, and F5P7) carried this diagnosis early in their disease course, although their muscle biopsies lacked primary inflammation, a hallmark of sIBM. Finger flexor weakness may be a rare manifestation of other inherited myopathies with various underlying genetic etiologies such as VCP and ACTA1,15 which were evaluated and ruled out by genetic testing. MYH7-related myopathy may present with proximal and distal weakness in the lower extremities with foot drop and toe drop as well as finger extensor weakness.16 However, while consistent for the toe involvement, the pattern of finger flexor weakness in MYH7-related myopathy (greater in digits IV and V than I, II, and III) is typically the reverse of what we observed in the patients in this series (greater in digits I, II, and III than IV and V). Thus, these unique clinical features help define the specific phenotype of ACTN2-related myopathy and help differentiate it from sIBM, and other hereditary distal myopathies.
MRI of the lower extremities in three patients in this series revealed a distinct pattern of asymmetric muscle involvement, with selective involvement of the hamstrings in the thigh, and the anterior tibial group and the soleus in the lower leg. This was particularly striking for the medial gastrocnemius in two patients, appearing nearly completely fat replaced on one side and unaffected on the other. This pattern was also seen in muscle imaging in one patient with biallelic c.1439A > G (p.Asn480Ser) ACTN2 variants9 but is distinct from patients with dominantly acting variants. Although selective involvement of the posterior and medial thigh compartments compared to anterior thigh is a common feature in limb-girdle muscular dystrophies, the selective involvement of the tibialis anterior, soleus, and medial gastrocnemius with marked side-to-side asymmetry distinguishes ACTN2 from some other hereditary myopathies. Recognition and characterization of patterns of muscle involvement is therefore a useful tool to aid in the diagnosis and variant interpretation of potential future patients with different biallelic ACTN2 variants.
Histologically, our series shows clear myopathic findings with marked disarray of the myofibrillar architecture, Type 1 fiber predominance and selective Type 1 fiber atrophy. Rimmed vacuolar pathology was consistently reported, similar to previous observations in both dominant and recessive ACTN2-related myopathy. Unlike previous reports, no clear core-like structures were identified on histology, or electron microscopy, which however was only available for F4P6 and F5P7. In the available EM images, areas of perinuclear and subsarcolemmal mitochondrial accumulation were notable, which typically lacked myofibrils. However, Z-disks were otherwise normal in appearance. Selective Type 1 fiber atrophy/hypotrophy can be a nonspecific histologic finding in myopathies. Of note, ACTN2 is expressed in Type 1 muscle fibers,17 and it remains unclear if these histological findings relate to a potential mechanistic connection between mutated ACTN2 and evolution of Type 1 fiber atrophy in muscle, thereby contributing to weakness. Given the small sample size, it is challenging to make a prediction on a possible correlation between muscle pathology and clinical severity specifically given the variability in the muscle biopsied and age of biopsy in our cohort, in addition to the inherent variability in muscle biopsies due to sampling bias.
ACTN2 is predominantly expressed in muscle and cardiac tissue. Currently, there is limited understanding between the various protein domains and the clinical manifestation of cardiac or skeletal disease, given that pathogenic variants are scattered throughout the ACTN2 gene without clear genotype–phenotype correlation. The (p.Arg506Gly) variant reported here, along with the previously reported recessive (p.Asn480Ser) variant, impact the spectrin repeat of ACTN2, which is essential for dimerization.9 Notably, dominantly acting pathogenic variants in the same exon have also been reported in patients with isolated cardiac disease (p.Tyr473Cys)18 and with isolated skeletal muscle disease (p.Cys487Arg).6 The lack of significant cardiac manifestations in our series is noteworthy. Overall, there seems to be a dichotomy where patients with predominant cardiac manifestations do not have concomitant progressive muscle disease, while patients presenting with skeletal myopathy do not show significant cardiac involvement, at least not early to mid-way in their disease progression. Although no dedicated screening was pursued, heterozygous carrier relatives in our series did not report cardiac or skeletal muscle involvement. Given the limited natural history data, regular long-term cardiac surveillance for all patients with ACTN2-related myopathy is essential.
The pathomechanism for ACTN2-related disease remains unclear and future mechanistic work is required to explore whether the ACTN2 Arg506Gly variant leads to impaired protein stability, altered localization, or impairment of ACTN2 function via another mechanism. The characterization of variants causative for cardiac disease revealed two distinct mechanisms in patient-derived iPSC-cardiomyocytes with a heterozygous truncated protein causing an arrhythmogenic cardiomyopathy, while in homozygosity manifesting as a severe progressive restrictive cardiomyopathy.19 A dominant-negative effect of the heterozygous dominant ACTN2 variant was postulated but has not been established for skeletal muscle variants.19 Haploinsufficiency has not yet been proven to be a disease mechanism in ACTN2-related disease although a handful of presumed heterozygous truncating variants have been reported as a cause for cardiac disease. The lower-than-expected number of loss of function variants in gnomAD (pLI score: 1, LOEUF of 0.24), indicates intolerance of ACTN2 to loss of function variation in a healthy control population. This is supported by knockdown approaches in animal models, ACTN2-null drosophila are lethal,20 and zebrafish show severe cardiac and skeletal muscle defects.21 The c.1516A>G ACTN2 variant identified in this series impacts the first base pair of exon 14 and has the potential to act as a complex hypomorph allele and to impair normal splicing. Unfortunately, we did not have frozen muscle tissue available to study the effect of the variant in disease relevant patient tissue. Therefore, we designed a splice vector assay, which did not show aberrant splicing in a cell-based system. This is in line with findings in homozygous (p.Asn480Ser) ACTN2 muscle in which ACTN2 expression level in patient muscle was similar to control.9 Taken together, these data suggest that recessive ACTN2 alleles do not act as a hypomorph, but that the missense variants interfere with the normal function of alpha-actinin 2. In patients with skeletal muscle disease, both dominant and recessive ACTN2 mutational mechanism result in progressive myopathy, of variable onset and progression, with histological findings of myofibrillar network disarray. Thus, there may be some overlapping concomitant effects on the sarcomere and muscle homeostasis in both mutational mechanisms, while the underlying alpha-actinin-2 dysfunction requires further investigations.
Given the phenotypic and genetic heterogeneity in skeletal and cardiac muscle disease variants, classification for ACTN2 remains challenging.22 This series is the first to report a clear genotype–phenotype correlation for (p.Arg506Gly) ACTN2, a recessive variant that is associated with a consistent and recognizable phenotype of adult-onset progressive proximal and lower extremity predominant myopathy with vacuolar histology and characteristic findings on muscle MRI imaging. Identification of additional variants and characterization of the spectrum of associated clinical, histological, and imaging findings is essential to gain further insight in the cardiac and skeletal muscle disease manifestations and potential genotype–phenotype correlations associated with pathogenic ACTN2 variants. Further work is needed to understand the underlying disease pathomechanisms in actinopathies, which now extends to include a recessive (p.Arg506Gly) ACTN2 variant.
Author Contributions
Study concept and design: SD, PM, AODL, CGB. Data acquisition: SD PM, MOL, DEB, TH, NA, AB, TM, MAS, DAD, JBS, SP, CAT, KH, JSC, KMM, JWC, AC, ARF, ALM. Data analysis and interpretation: SD PM, MOL, DEB, TH, NA, AB, TM, MAS, DAD, JBS, SP, CAT, KH, JSC, KMM, JWC, AC, ARF, ALM, MTW, AODL, CGB. Drafting the manuscript: SD, PM, CGB. Study supervision: MTW, AODL, CGB. Critical review of the manuscript: All authors.
Acknowledgments
We thank the families for participating in this study, and Christopher Mendoza, Gilberto (“Mike”) Averion and Kia Brooks for their help in the clinic. We also like to thank Kyle Satterstrom for his help with AlphaMissense. The work in C.G. Bönnemann's laboratory is supported by intramural funds from the NIH National Institute of Neurological Disorders and Stroke. Sequencing and analysis were provided by the Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG) and was funded by the National Human Genome Research Institute (NHGRI), the National Eye Institute, the National Heart, Lung and Blood Institute grant UM1HG008900 to Daniel MacArthur, Heidi Rehm, and Anne O'Donnell-Luria, and NHGRI grant R01 HG009141 and grant 2020-224274 from the Chan Zuckerberg Initiative DAF, an advised fund of the Silicon Valley Community Foundation, to the Rare Genomes Project. Sequencing and analysis for Individual F3P5 was performed by the Care4Rare-Solve Canada, funded by Genome Canada, the Canadian Institutes of Health Research, the Ontario Genomics Institute, Ontario Research Fund, Genome Quebec, and Children's Hospital of Eastern Ontario Foundation. Clinical evaluation, sequencing, and analysis for Individual F5P7 was performed by the Undiagnosed Diseases Network, funded by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director under Award Number U01HG010218. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Members of the Undiagnosed Diseases Network: Maria T. Acosta, David R. Adams, Raquel L. Alvarez, Justin Alvey, Aimee Allworth, Ashley Andrews, Euan A. Ashley, Ben Afzali, Carlos A. Bacino, Guney Bademci, Ashok Balasubramanyam, Dustin Baldridge, Jim Bale, Michael Bamshad, Deborah Barbouth, Pinar Bayrak-Toydemir, Anita Beck, Alan H. Beggs, Edward Behrens, Gill Bejerano, Hugo J. Bellen, Jimmy Bennett, Jonathan A. Bernstein, Gerard T. Berry, Anna Bican, Stephanie Bivona, Elizabeth Blue, John Bohnsack, Devon Bonner, Lorenzo Botto, Lauren C. Briere, Gabrielle Brown, Elizabeth A. Burke, Lindsay C. Burrage, Manish J. Butte, Peter Byers, William E. Byrd, John Carey, Thomas Cassini, Sirisak Chanprasert, Hsiao-Tuan Chao, Ivan Chinn, Gary D. Clark, Terra R. Coakley, Laurel A. Cobban, Joy D. Cogan, Matthew Coggins, F. Sessions Cole, Heather A. Colley, Rosario Corona, William J. Craigen, Andrew B. Crouse, Michael Cunningham, Precilla D'Souza, Hongzheng Dai, Surendra Dasari, Joie Davis, Jyoti G. Dayal, Margaret Delgado, Esteban C. Dell'Angelica, Katrina Dipple, Daniel Doherty, Naghmeh Dorrani, Argenia L. Doss, Emilie D. Douine, Dawn Earl, David J. Eckstein, Lisa T. Emrick, Christine M. Eng, Marni Falk, Elizabeth L. Fieg, Paul G. Fisher, Brent L. Fogel, Jiayu Fu, William A. Gahl, Ian Glass, Page C. Goddard, Rena A. Godfrey, Andrea Gropman, Meghan C. Halley, Rizwan Hamid, Neal Hanchard, Kelly Hassey, Nichole Hayes, Frances High, Anne Hing, Fuki M. Hisama, Ingrid A. Holm, Jason Hom, Martha Horike-Pyne, Alden Huang, Yan Huang, Sarah Hutchison, Wendy Introne, Kosuke Izumi, Gail P. Jarvik, Jeffrey Jarvik, Suman Jayadev, Orpa Jean-Marie, Vaidehi Jobanputra, Emerald Kaitryn, Shamika Ketkar, Dana Kiley, Gonench Kilich, Shilpa N. Kobren, Isaac S. Kohane, Jennefer N. Kohler, Susan Korrick, Deborah Krakow, Donna M. Krasnewich, Elijah Kravets, Seema R. Lalani, Christina Lam, Brendan C. Lanpher, Ian R. Lanza, Kimberly LeBlanc, Brendan H. Lee, Richard A. Lewis, Pengfei Liu, Nicola Longo, Sandra K. Loo, Joseph Loscalzo, Richard L. Maas, Ellen F. Macnamara, Calum A. MacRae, Valerie V. Maduro, AudreyStephannie Maghiro, Rachel Mahoney, May Christine V. Malicdan, Laura A. Mamounas, Teri A. Manolio, Rong Mao, Ronit Marom, Gabor Marth, Beth A. Martin, Martin G. Martin, Julian A. Martínez-Agosto, Shruti Marwaha, Allyn McConkie-Rosell, Alexa T. McCray, Elisabeth McGee, Matthew Might, Mohamad Mikati, Danny Miller, Ghayda Mirzaa, Eva Morava, Paolo Moretti, Marie Morimoto, John J. Mulvihill, Mariko Nakano-Okuno, Stanley F. Nelson, Shirley Nieves-Rodriguez, Donna Novacic, Devin Oglesbee, James P. Orengo, Laura Pace, Stephen Pak, J. Carl Pallais, Jeanette C. Papp, Neil H. Parker, Leoyklang Petcharet, John A. Phillips III, Jennifer E. Posey, Lorraine Potocki, Barbara N. Pusey Swerdzewski, Aaron Quinlan, Deepak A. Rao, Anna Raper, Wendy Raskind, Genecee Renteria, Chloe M. Reuter, Lynette Rives, Amy K. Robertson, Lance H. Rodan, Jill A. Rosenfeld, Elizabeth Rosenthal, Francis Rossignol, Maura Ruzhnikov, Marla Sabaii, Jacinda B. Sampson, Timothy Schedl, Kelly Schoch, Daryl A. Scott, Elaine Seto, Prashant Sharma, Vandana Shashi, Emily Shelkowitz, Sam Sheppeard, Jimann Shin, Edwin K. Silverman, Janet S. Sinsheimer, Kathy Sisco, Kevin S. Smith, Lilianna Solnica-Krezel, Ben Solomon, Rebecca C. Spillmann, Andrew Stergachis, Joan M. Stoler, Kathleen Sullivan, Shirley Sutton, David A. Sweetser, Virginia Sybert, Holly K. Tabor, Queenie K.-G. Tan, Amelia L. M. Tan, Arjun Tarakad, Herman Taylor, Mustafa Tekin, Willa Thorson, Cynthia J. Tifft, Camilo Toro, Alyssa A. Tran, Rachel A. Ungar, Tiina K. Urv, Adeline Vanderver, Matt Velinder, Dave Viskochil, Tiphanie P. Vogel, Colleen E. Wahl, Melissa Walker, Nicole M. Walley, Jennifer Wambach, Jijun Wan, Lee-kai Wang, Michael F. Wangler, Patricia A. Ward, Daniel Wegner, Monika Weisz Hubshman, Mark Wener, Tara Wenger, Monte Westerfield, Matthew T. Wheeler, Jordan Whitlock, Lynne A. Wolfe, Kim Worley, Shinya Yamamoto, Zhe Zhang, Stephan Zuchner.
Conflict of Interest
KMc and AC are an employee of GeneDx, LLC. The remaining authors declare that they have no conflict of interest.




