Short-chain fatty acid producers in the gut are associated with pediatric multiple sclerosis onset
Abstract
Objective
The relationship between multiple sclerosis and the gut microbiome has been supported by animal models in which commensal microbes are required for the development of experimental autoimmune encephalomyelitis. However, observational study findings in humans have only occasionally converged when comparing multiple sclerosis cases and controls which may in part reflect confounding by comorbidities and disease duration. The study of microbiome in pediatric-onset multiple sclerosis offers unique opportunities as it is closer to biological disease onset and minimizes confounding by comorbidities and environmental exposures.
Methods
A multicenter case–control study in which 35 pediatric-onset multiple sclerosis cases were 1:1 matched to healthy controls on age, sex, self-reported race, ethnicity, and recruiting site. Linear mixed effects models, weighted correlation network analyses, and PICRUSt2 were used to identify microbial co-occurrence networks and for predicting functional abundances based on marker gene sequences.
Results
Two microbial co-occurrence networks (one reaching significance after adjustment for multiple comparisons; q < 0.2) were identified, suggesting interdependent bacterial taxa that exhibited association with disease status. Both networks indicated a potentially protective effect of higher relative abundance of bacteria observed in these clusters. Functional predictions from the significant network suggested a contribution of short-chain fatty acid producers through anaerobic fermentation pathways in healthy controls. Consistent family-level findings from an independent Canadian-US study (19 case/control pairs) included Ruminococaccaeae and Lachnospiraceae (p < 0.05). Macronutrient intake was not significantly different between cases and controls, minimizing the potential for dietary confounding.
Interpretation
Our results suggest that short-chain fatty acid producers may be important contributors to multiple sclerosis onset.
Introduction
Multiple sclerosis (MS) affects over 2.8 million people worldwide, resulting in chronic disability and high socioeconomic burden.1, 2 The risk of developing MS is thought to be driven by genetic and environmental factors.3-5 Recently, the gut microbiome has been proposed as not only a mediator of the effect of known environmental risk factors but also as an independent risk factor for MS.6 This relationship has been supported by animal models where commensal microbiota were required for the development of experimental autoimmune encephalomyelitis.7
Observational studies typically have not identified differences in gut microbiome diversity when comparing MS cases and controls.8, 9 Despite occasional convergence in findings suggesting individual taxa differing in their relative abundance, there is still a considerable amount of variability between studies that may reflect confounding.8, 10 The instrinsic interindividual heterogeneity of healthy gut microbiome composition is further broadened by differences in human lifestyle and physiological factors, limiting our ability to identify causal relationships.10 Studying pediatric-onset MS (POMS), which may be diagnosed closer to the biological onset of the disease, potentially minimizes the effect of comorbidities and environmental exposures, offering a unique opportunity to unravel the microbiome's contribution to MS pathogenesis.9
Therefore, we aimed to characterize the bacterial taxonomic profile of POMS patients in a multicenter, matched case–control study utilizing 16S rRNA gene sequencing of stool samples collected shortly after MS symptom onset and replicate findings in a second dataset.
Methods
Study population
Individuals with MS onset before age 18 years and healthy controls were recruited from seven sites in the U.S. Network of Pediatric MS Centers including the University of California San Francisco, State University of New York at Buffalo, University of Alabama at Birmingham, Boston Children's Hospital, Stony Brook University Medical Center, Children's Hospital of Philadelphia, and New York University between 2012 and 2018. Healthy controls attended general pediatric clinics at the participating institutions and did not have a personal or familial history of autoimmune disorders. POMS cases were within 5 years of symptom onset and met the 2010 McDonald criteria for MS.11 All participants were tested for the presence of myelin oligodendrocyte glycoprotein antibodies at the Mayo Clinic neuroimmunology laboratory (Rochester, MN) using a live cell-based flow cytometry assay. Those with positive results were excluded and cases with low titers had their final diagnosis ascertained by two MS experts, as previously described.12 Additionally, individuals exposed to systemic antibiotics, probiotics, or steroids within 30 days before sample collection were excluded. POMS cases on a disease-modifying therapy (DMT) were invited to enroll if on stable dosing for more than three months. POMS patients were 1:1 matched with healthy controls by their age at stool sample collection (±3 years), sex, self-reported race, ethnicity, and recruiting site. Institutional Review Board approval was obtained from all participating institutions. Participants and their parent or legal guardian provided, respectively, assent and informed consent.
Gut microbiota profiling
The participant's first stool of the day was collected by a parent and shipped overnight on ice to the University of California, San Francisco and stored at −80°C before processing. DNA was extracted, and the V4 region of the bacteria 16S ribosomal RNA (rRNA) gene was amplified for sequencing as previously described.13 16S rRNA sequencing data were processed as previously described.14 Forward and reverse reads were processed separately and quality filtered using the DADA2 package version 1.9.0.15 in R, version 4.2.1 (the R Foundation). Reads with more than two expected errors were removed and forward and reverse reads were confirmed to be of at least 150 base pairs in length. Error rates of the filtered and dereplicated reads were estimated using 100,000 sequences. Paired end reads with a minimum overlap of 25 base pairs were merged to obtain the full denoised sequences. Chimeras and any abnormally short or long sequences (±5 bases from the median number of reads) were removed. Amplicon sequence variants (ASVs), which permit analysis of 16S RNA variants that differ by as little as one nucleotide, were used for downstream analysis. Taxonomy was assigned using the naive Bayesian classifier method (Kingdom to Family) and exact string matching (Genus and Species) utilized the SILVA v132 reference database.15-17 It is important to note that while an ASV has a unique nucleotide sequence, it might not be assigned a unique species due to the limited capacity of short-read 16S rRNA sequencing to differentiate phylogenetically related species or strains. Using the decontam package with previously described parameters,18 ASVs with a contaminant classification threshold p < 0.1 were removed.19 ASVs containing <1/1000th of a percent of total reads were removed. Sequencing reads were representatively rarefied to the minimum sequencing depth (84,818 reads/sample) 100 times, and the rarefied sample profile closest to the sample-specific centroid was selected, as described previously.13 The resulting tables included 1482 ASVs.
Covariates
Participants' basic demographics and clinical history were collected with standardized forms completed by the parent or legal guardian and assisted by a research coordinator. Patients with POMS had their MS-related data extracted from a prospective pediatric MS registry. The season at stool sample collection variable was defined as winter (December, January, and February), spring (March, April, and May), summer (June, July, and August), and autumn (September, October, and November). Exposure to breastfeeding was deemed present if the individual was ever breastfed, irrespective of duration. The DMT variable was defined as treatment naive versus ever exposed. Recent intake of foods and nutrients was standardized for all participants based on their responses to the Block Kids Food Screener, a food frequency questionnaire (FFQ) developed by NutritionQuest.20 All covariates were measured at stool sample collection (i.e., study baseline), except for body mass index (BMI) and the expanded disability status scale (EDSS), which were measured at the visit date closest to when the stool sample was received.
Microbial diversity
For microbial diversity analysis, α-diversity was measured by Shannon,21 Chao1,22 and Faith's phylogenic diversity indexes. β-diversity was assessed using Weighted UniFrac23 distances and visualized using principal coordinates analysis (PCoA). The referred analyses were performed with QIIME2.24 A linear mixed effects model was applied to estimate the difference of α-diversity indexes between individuals with POMS and healthy controls, adjusting for a priori-defined fixed effects of age (continuous), BMI (continuous), and sex and random effects of recruiting site, season, and matching pairs. The PERMANOVA test25 was used to assess the effect of host confounding factors on the variation of microbiome abundance and was performed by using the “adonis2” function implemented in R package vegan.26 Weighted UniFrac distances were compared individually for each reported host factors (disease status (cases/controls), site, age, race, ethnicity, sex, BMI, DMT usage, breastfeeding, birth delivery mode, season at sample collection, macronutrients consumption, HLA-DRB1 genotype, serum vitamin D levels, Epstein–Barr virus (viral capsid antigen) (EBV-VCA), herpes simplex virus type 1 (HSV1), and cytomegalovirus (CMV) serostatus). The variance of microbial abundance between POMS and controls was estimated after adjusting for sample collecting site, season, and matching pairs. The empirical p-value was obtained by running 999 permutations.
Microbial composition and disease status
Microbial composition was normalized as relative abundance and further transformed using an arcsine square root transformation.27-29 Linear mixed effects models were applied to the transformed data to identify ASVs associated with disease status. The model had ASV abundance as its outcome and was adjusted by fixed effects of breastfeeding (after assessing the effect of host confounding factors in PERMANOVA analyses) and a priori-defined random effects of recruiting site, season, and matching pairs. The fixed effects were reassessed after determining the effect of host confounding factors on the variance of microbiome abundance (Adonis R2). The inclusion of season at sample collection as a random effect was done a priori to account for expected variations in dietary habits that could have influenced microbiome composition. Additionally, stratification by treatment status was performed using separate models. Linear regression was performed using lmer function from R package “lme4.” To reduce the effect of zero-inflation in microbiome data, a variance filtering step was applied to remove species features with very low variance (lower than the median variance).
Microbial co-abundance network
Co-abundance network inference of ASVs present in at least 20% of samples (resulting in 268 ASVs) was performed using the SparCC method in R using the SpiecEasi package.30, 31 Microbial modules were identified by weighted correlation network analysis (WGCNA) as described.14, 32 Briefly, the SparCC correlation matrix was generated and transformed to an adjacency matrix using soft thresholding, and a topology overlap matrix was generated. The topology overlap matrix was hierarchically clustered using an average linkage in hclust. The resulting dendrogram was cut using dynamicTreeCut in the stats package to generate clusters with at least three ASVs. Correlated modules (r ≥ 0.5) were combined, generating a dissimilarity matrix for further hierarchical clustering. The quantitative values of each module were calculated for each participant from module eigengenes, defined as the first principal component of the abundance matrix of a respective module. Each module eigengene was examined for its association with disease status using linear mixed effects models adjusting for fixed effect of breastfeeding and random effects of recruiting site, season, and matching pairs.
Metagenomic prediction
Metagenomic functional profiling of conserved 16S rRNA from microbial communities was predicted using PICRUSt2.33 Significant modules identified from the microbial co-abundance network analysis were grouped into MetaCyc metabolic pathways.33 Pathway abundances were dichotomized by their respective median abundance. The association between the predicted metabolic pathways and the disease status was estimated by conditional logistic regression models adjusted by the a priori-defined set of covariates BMI and season, and conditioned on matching pairs using the survival package. The predicted metabolic pathways are thought to be mediators of the association between co-abundance networks and disease status, thus a different set of confounders for this relationship was a priori defined and independent from the set adapted after the PERMANOVA test findings.34
Diet analysis
Four dietary components (carbohydrate, protein, fiber and fat) were extracted from the FFQ, and the amount of each dietary component consumed in grams was compared between cases and controls with paired t-tests. Associations between each dietary component and each ASVs were estimated using linear mixed effects models adjusted for the a priori-defined fixed effects of age and BMI, as well as random effects of recruiting site, season, and matching pairs.
Complementary analysis
An external dataset comprised of POMS cases and healthy controls recruited from four Canadian and one American site was used for assessing the reproducibility of findings. POMS were 1:1 matched on sex, self-identified race (grouped as white/other/missing), site (Canada/United States), and age (±3 years) at sample collection. Gut microbiome profiling was performed as previously described.9 Analytical procedures were replicated as described under the sections “Microbial composition and disease status” and “Microbial co-abundance network,” except for season and breastfeeding adjustment since variables were not available. The linear mixed effect models were adjusted for random effects of site and matching pairs. As per privacy and data sharing agreements, these analyses were conducted in Canada.
Statistical considerations
All statistical analyses were conducted using two-sided tests and the alpha level was set at 0.05, unless otherwise specified. Tests of microbial composition, microbiome networks or metagenomic predictions and disease status or food intake were controlled with Benjamini–Hochberg's false discovery rate (FDR).35 Given the limited sample size and expected high interindividual variability, a less conservative FDR threshold of 0.2 was used. Results are presented as coefficients when estimated by linear mixed effects models and odds ratios (OR) when estimated by conditional logistic regression alongside their respective 95% confidence intervals (CI).
Results
Stool samples from 60 individuals with POMS and 43 healthy individuals were obtained. The matching procedure identified 35 pairs including 24 exact (age ±3 years, with same sex, self-reported race, ethnicity, and recruiting site) and 11 close matches (age ±3 years, different site, race, and/or sex). Baseline characteristics stratified by disease status are outlined in Table 1. Cases and controls presented a similar proportion of females (77.1% vs. 74.3%), white race (71.4% vs. 80%), Hispanic/Latino ethnicity (28.6% vs. 28.6%), and median age at stool sample collection (15.7 vs. 16.1 years). POMS cases had a median disease duration of 0.9 years (IQR 0.3–1.4) and 54% were on stable DMT. POMS cases that were on DMTs at baseline had similar median age (14.4 vs. 14.2 years) and disease duration (0.86 vs. 0.87 years) as DMT-naive POMS.
| Characteristics | POMS (n = 35) | Control (n = 35) |
|---|---|---|
| Sex (female), n (%) | 27 (77.1%) | 26 (74.3%) |
| Race (White), n (%) | 25 (71.4%) | 28 (80%) |
| Ethnicity (Hispanic or Latino), n (%) | 10 (28.6%) | 10 (28.6%) |
| Age (years), median (IQR) | 15.9 (14.5, 16.8) | 16.0 (15.1, 17.5) |
| Body mass index, median (IQR) | 23.8 (20.6, 30.1) | 22.4 (20.1, 28.3) |
| EDSS, median (IQR) | 1.5 (0, 1.5) | |
| Disease duration (years), median (IQR) | 0.9 (0.3, 1.4) | |
| DMT usage (yes), n (%) | 19 (54%) | – |
| Breastfed, n (%) | ||
| Yes | 24 (68.6%) | 27 (77.1%) |
| No | 6 (17.1%) | 8 (22.9%) |
| Unknown | 5 (14.3%) | 0 |
| Birth delivery mode, n (%) | ||
| Vaginal | 28 (80%) | 27 (77.1%) |
| Cesarean section | 6 (17.1%) | 4 (11.4%) |
| Unknown | 1 (2.9%) | 4 (11.4%) |
- Disease modifying therapies (DMTs) included glatiramer acetate (Copaxone, n = 12), interferon beta-1a (Avonex, n = 2), interferon beta-1a (Rebif, n = 1), interferon beta-1b (Betaseron, n = 1), natalizumab (Tysabri, n = 1), peginterferon beta-1a (Plegridy, n = 1).
- EDSS, Expanded Disability Status Scale; IQR, interquartile range.
Microbial diversity
α-diversity, or within-individual diversity, was measured at both the species and phylogenetic level. Species diversity, the total number of different species, measured by the Shannon index and Chao1, was not significantly different between POMS individuals and controls (respectively, p = 0.23 and p = 0.08). However, using phylogenetic-based distance measures, that quantify the evolutionary relatedness of groups of microbes within a community, the mean diversity became significantly different when measured by Faith's PD index (p = 0.02). POMS cases presented consistently lower levels of phylogenetic diversity than controls after adjustment for potential confounders, study design, and clustering by site and season of sample collection (Fig. 1A).
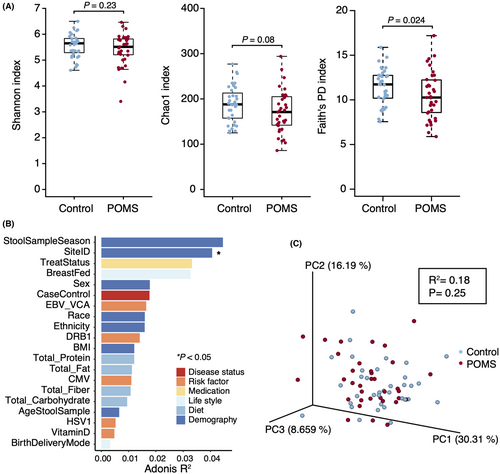
The effect of host confounding factors on the variance of microbiome abundance was significantly different for the study site and larger, but not significantly different, for season of sample collection, use of DMTs and history of breastfeeding (Fig. 1B). This observation prompted the inclusion of breastfeeding as a fixed effect for model adjustment as opposed to BMI when estimating the association between microbial diversity or co-abundance network and disease status.
β-diversity, or between-individual diversity, using a phylogenetic distance-based measure was not significantly different between POMS cases and controls after adjustment (p = 0.25) and no clustering patterns are observed in the PCoA (Fig. 1C).
Microbial composition and disease status
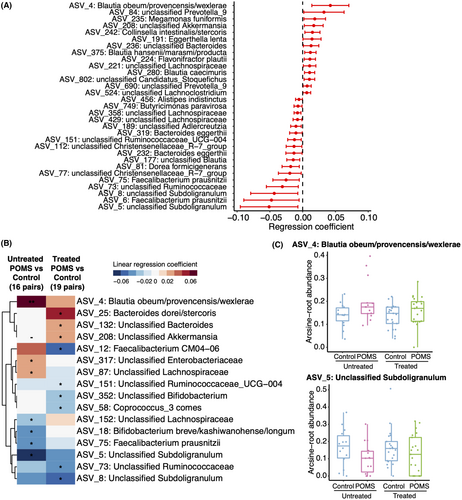
Among the 1482 ASVs identified, 31 were associated with disease status after adjusting for confounders and are presented in Figure 2A (p < 0.05). However, none remained significant after adjustment for multiple comparisons. Taxa with regression coefficients greater than zero such as those observed in Blautia obeum/provencis/wexlerae (ASV 4) and Prevotella (ASV 84) indicate higher abundance of those taxa in POMS cases while those with coefficients lower than zero, such as Subdoligranulum (ASV 5, ASV 8), represent a lower abundance in POMS cases relative to controls. After stratifying the models by POMS treatment status, Blautia (ASV 4), Faecalibacterium prausnitzii (ASV 75), and Subdoligranulum (ASV 5) remained significantly associated with disease status but only for those that were DMT-naive (p < 0.05, Fig. 2B). Of note, Blautia (ASV 4) presented a higher mean relative abundance in DMT-naive POMS (n = 16) when compared to controls (p < 0.01, Fig. 2B,C). The opposite was observed for Subdoligranulum (ASV 5), where a lower relative abundance was observed in DMT-naive POMS relative to controls (p < 0.05, Fig. 2B,C). Despite presenting a less pronounced difference, the direction of the association was persistent for the difference in relative abundance of Blautia (ASV 4), Faecalibacterium prausnitzii (ASV 75), and Subdoligranulum (ASV 5) between POMS exposed to DMTs and controls. Akkermansia (ASV 208) presented higher relative abundance in POMS that was driven by patients on DMTs. Due to the low variance of Akkermansia (ASV 208) among DMT-naive cases and controls, this genus was filtered (reducing the effect of zero-inflation, as a priori defined) and no estimates were provided for this subgroup.
Microbial co-abundance network
WGCNA clustering of the SparCC correlation matrix generated 17 modules of ASVs for co-abundance analysis. Those modules aim to identify dependencies between species, suggesting natural microbial communities. Each was tested for association with disease status; two were significantly associated with disease status, although only one remained significant after multiple comparisons adjustment. In both networks, POMS cases had lower abundances of the microbes in each cluster: pink (p < 0.05, q < 0.2) and purple (p < 0.05, q = 0.22). The clusters comprised 18 ASVs, some of which were identified above to be individually associated with disease status. Of note, Subdoligranulum (ASV 5, ASV 8) was present in one of the significant modules (pink), while Blautia (ASV 177) was in the other (purple) (Table 2, modules were arbitrarily identified using color names and do not reflect colors used in figures).
| Phylum | Class | Order | Family | Genus | Species | Signa | Moduleb | |
|---|---|---|---|---|---|---|---|---|
| ASV_151 | Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Ruminococcaceae_UCG-004 | NA | 1 | Pink |
| ASV_229 | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Roseburia | Hominis | 0 | Pink |
| ASV_24 | Firmicutes | Erysipelotrichia | Erysipelotrichales | Erysipelotrichaceae | Erysipelotrichaceae_UCG-003 | NA | 0 | Pink |
| ASV_321 | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Lachnospiraceae_FCS020_group | NA | 0 | Pink |
| ASV_40 | Bacteroidetes | Bacteroidia | Bacteroidales | Rikenellaceae | Alistipes | Putredinis | 0 | Pink |
| ASV_5 | Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Subdoligranulum | NA | 1 | Pink |
| ASV_58 | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Coprococcus_3 | Comes | 0 | Pink |
| ASV_60 | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | NA | NA | 0 | Pink |
| ASV_62 | Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Ruminiclostridium_5 | NA | 0 | Pink |
| ASV_8 | Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Subdoligranulum | NA | 1 | Pink |
| ASV_104 | Proteobacteria | Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | Bilophila | Wadsworthia | 0 | Purple |
| ASV_122 | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Coprococcus_1 | Catus | 0 | Purple |
| ASV_13 | Actinobacteria | Coriobacteriia | Coriobacteriales | Coriobacteriaceae | Collinsella | Aerofaciens | 0 | Purple |
| ASV_177 | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Blautia | NA | 1 | Purple |
| ASV_22 | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Dorea | Longicatena | 0 | Purple |
| ASV_269 | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Blautia | NA | 0 | Purple |
| ASV_520 | Actinobacteria | Coriobacteriia | Coriobacteriales | Coriobacteriales_Incertae_Sedis | NA | NA | 0 | Purple |
| ASV_81 | Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Dorea | formicigenerans | 1 | Purple |
- a Indicates whether the ASVs are significantly different (p < 0.05, bolded) between POMS cases and controls in single ASV analysis (1 different, 0 not different).
- b Microbial co-abundance network modules were identified using arbitrary color names and do not reflect colors used in figures.
Metagenomic functional pathways prediction
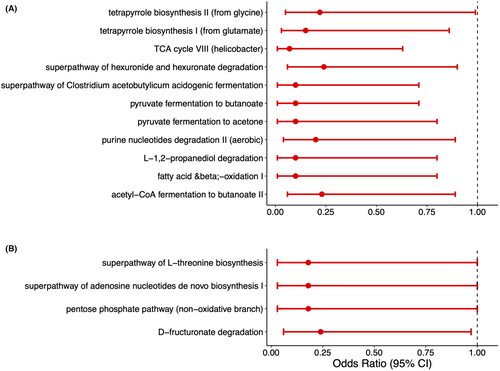
The metabolic pathways predicted from the significant gut microbial co-abundance network modules (pink and purple) are depicted in Figure 3. Of the 150 predicted pathways for the ASVs from the pink module, 11 were significantly associated with disease status (p < 0.05, Fig. 3A). All 11 had a higher pathway abundance among controls than POMS cases. The largest effect sizes (point estimates farther away from the null value of 1) were observed in pathways associated with anaerobic fermentation leading to the production of short-chain fatty acids (SCFA) and lower odds of having POMS. Namely, pyruvate fermentation to butanoate (OR: 0.10 95% CI 0.01–0.71, p = 0.02), pyruvate fermentation to acetone (OR: 0.10 95% CI 0.01–0.80, p = 0.03), superpathway of Clostridium acetobutylicum acidogenic fermentation (OR: 0.10 95% CI 0.01–0.71, p = 0.02), L-1,2-propanediol degradation (OR: 0.10 95% CI 0.01–0.80, p = 0.03), and acetyl-CoA fermentation to butanoate (OR: 0.23 95% CI 0.06–0.89, p = 0.03). Of the 165 predicted pathways for the purple module, four were marginally significantly associated with POMS status (p < 0.05, Fig. 3B).
Diet analysis
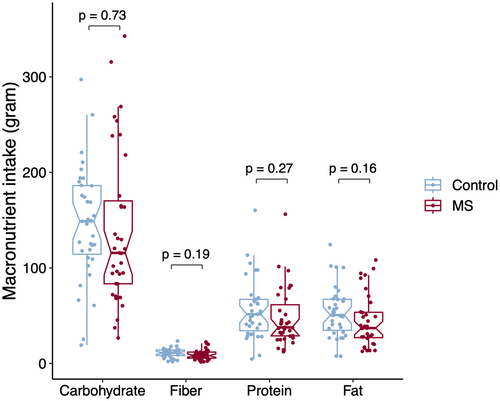
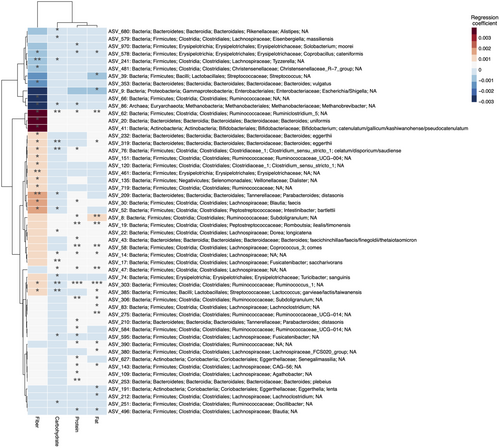
The mean carbohydrate, fiber, protein, and fat intake was not significantly different between POMS and controls (Fig. 4). The ASVs with relative abundance significantly associated with different levels of macronutrients intake (p < 0.05, q > 0.2) are represented in Figure 5. The larger effects on relative abundance were associated with higher fiber intake driving lower relative abundances of Ruminococcaceae (ASV 66) and Methanobrevibacter (ASV 86) and higher abundances of Ruminoclostridium (ASV 62), Bacterioides uniformis (ASV 20), and Bifidobacterium (ASV 41). In addition to Ruminoclostridium (ASV 62), Ruminococcaceae (ASV 151) also belonged to the pink module and was associated with fiber intake levels, but with a less pronounced difference. Further ASVs associated with diet in the pink module included Subdoligranulum (ASV 8) and Coprococcus comes (ASV 58) both had higher relative abundances with higher fat and protein intake. From the purple module, only Dorea longicatena (ASV 22) abundance was associated with diet, slightly increasing with carbohydrate consumption.
Complementary analysis
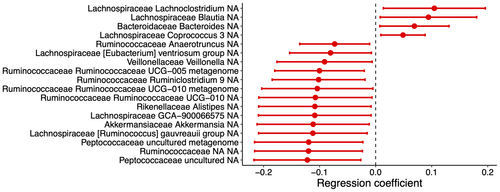
The matching procedure identified 19 pairs of POMS and healthy controls in the replication dataset. Cases and controls presented the same proportion of females (78.9%), White race (42.1%), site origin (84.2% from Canada), and similar median age at stool sample collection (17.6 vs. 15.5 years) (Table 3). ASVs associated with disease status after adjustment for potential confounders are presented in Figure 6 (p < 0.05). None remained significant after multiple comparisons adjustment. Multiple ASVs from the Ruminococcaceae family (UCG-010, UCG-005, Ruminiclostridium 9, and Anaerotruncus) present a lower abundance in POMS cases relative to controls. The same was observed for three ASVs from the Lachnospiraceae family (GCA-900066575, Ruminococcus gauvreauii, and Eubacterium ventriosum) while three ASVs from this family presented a higher abundance (Lachnoclostridium, Coprococcus 3, and Blautia). Akkermansia presented lower relative abundance in POMS. The WGCNA clustering procedure yielded 31 modules of ASVs, none being associated with disease status (data not shown).
| Characteristics | POMS (n = 19) | Control (n = 19) |
|---|---|---|
| Sex (female), n (%) | 15 (78.9%) | 15 (78.9%) |
| Race (Whitea), n (%) | 8 (42.1%) | 8 (42.1%) |
| Age (median, IQR) | 17.6 (16.3, 18.3) | 15.5 (14.9, 16.3) |
| Disease duration (years), median (IQR) | 0.7 (0.4, 4.0) | – |
| Site (Canada), n (%) | 16 (84.2%) | 16 (84.2%) |
- IQR, interquartile range; NA, not applicable.
- a Three subjects were matched on race at the missing category.
Discussion
In this matched case–control study, POMS individuals with recent disease onset had different relative abundances of several gut bacteria in comparison to healthy controls. Stratified analysis by DMT status preserved the directionality of the association of most ASVs with disease status and had minor influence on their effect magnitude. There is no evidence that these results were affected by variation in disease duration or age at stool sample collection when comparing DMT treated to untreated POMS patients. The two microbial co-occurrence networks identified by WGCNA, suggesting interdependent bacterial taxa, exhibited an association with disease status, one after adjustment for multiple comparisons. This finding, in turn, suggested a potentially protective effect of having more of the bacteria observed in these clusters. The metagenomic predictions from the significant network (pink) suggested a prominent contribution of SCFA production through anaerobic fermentation pathways. Among these known SCFA producers, bacteria from the Ruminococcaceae family were consistently observed in the main and complementary analyses. SCFA are primarily produced in the proximal colon of healthy subjects by bacterial fermentation of nondigestible carbohydrates and have been associated with anti-inflammatory properties.36
In past studies, microbial diversity, particularly α-diversity, has been frequently reported as not significantly different between MS cases and controls.8, 37 In the present study, only one measure of α-diversity that incorporated phylogenetically derived distances was significantly lower among POMS cases compared to controls. Phylogenetic diversity is expected to predict functional similarity, given that species more closely related are morphologically similar and assume similar functional roles in their respective ecosystems.38 Our careful matching procedure and the age range of our cohort may have allowed for uncovering this finding, as a prior study in adult MS discordant twins found no significant differences.39 Overall, a lower α-diversity characterizes a less healthy gut microbiota community.40
Given the modest sample size in POMS studies, ASVs associated with disease status that presented large effect sizes but did not reach the multiple comparisons threshold were reported in this study. Among those, a lower relative abundance of taxa from the Lachnospiraceae (unclassified genera) and Ruminococcaceae families (Faecalibacterium prausnitzii, Subdoligranulum, and UCG-004) were observed in POMS cases. These families are known for their important contribution to butyrate and propionate production from carbohydrate fermentation.41 Our complementary analysis observed a similar and consistent directionality of effect for significant ASVs from the Ruminococcaceae family. Diminished relative abundances from one taxa of Lachnospiraceae relative to healthy controls has only been previously reported by the Canadian group.9 Our complementary analysis has expanded this finding to six significant taxa from this family, although distinct abundance patterns were observed. More consistently, a lower relative abundance of Faecalibacterium prausnitzii has been reported in case–control studies in relapsing–remitting MS.8, 37, 42 Of note, their reduced abundance of this species has also been frequently reported in other immune-mediated disorders such as Crohn's disease.43 Similarly to another Canadian study, a lower abundance of Subdoligranulum in individuals with MS compared to healthy controls was found in the present study but was not reproduced until now.44 Consistent with prior studies, Akkermansia, a mucin-degrading bacteria capable of producing acetate and propionate, was found at a higher relative abundance in POMS than controls.8, 37 Nevertheless, this finding was not significant when comparing the subset of DMT-naive POMS to controls, which, alongside other subgroup analyses, should be interpreted with caution due to the limited sample size. Additionally, a lower relative abundance of Akkermansia in POMS than controls was observed in the complementary analysis. In contrast to a study from the International Multiple Sclerosis Microbiome Study consortium, we found Blautia had a higher relative abundance in POMS relative to controls.37 Notably, the 1152 subject consortium study enrolled older adults and used household controls (averaging 50.6 years) which, despite being well matched on the intended covariates, resulted in highly sex-discordant pairs, thereby limiting comparability.10 In line with our findings, which were further reproduced in the complementary analysis, is a study of adults that found higher abundances in 31 MS patients and 36 age and sex-matched healthy controls.45 However, comparisons were restricted to the genus level, and different species could exert different effects. Regardless of belonging to the colonic dominant phylum Firmicutes, which is composed of many SCFA-producing species, Blautia cannot produce butyrate from carbohydrates.41
Microbial communities are expected to naturally occur and share functional dependencies. In this study, we have identified clusters of bacteria from the Ruminococcaceae and Lachnospiraceae families associated with disease status that contained some of the individual ASVs highlighted above in the main (species level) and complementary (family level) analysis. Among the shared predicted metabolic pathways expressed by those microbes, more abundant and exerting a larger effect were those leading to the production of SCFA. In preclinical studies, SCFAs have been shown to exert anti-inflammatory effects modulating the systemic immune response at the gut barrier level positively affecting the experimental autoimmune encephalomyelitis disease course.36 As PICRUSt only predicts conserved functional traits and cannot distinguish strain-specific functionality,33 these data should be viewed mostly as hypothesis-generating.46 However, in an open label non-randomized trial, propionic acid was administered to 97 MS patients for 2 weeks before DMT initiation, and findings suggested a lesser disease progression compared to 57 historical MS controls.47 Baseline levels of propionic acid were lower in serum and stool samples of MS patients compared to healthy controls and baseline regulatory T cells were also lower in MS patients.47
Diet can be an important source of interindividual heterogeneity in the gut microbiota profile.10 All participants filled out a pre-validated FFQ, and the mean intake of macronutrients was similar between cases and controls. Thus, while diet was associated with differences in some taxa, diet was less likely to explain the differences in the gut microbiota between our cases and controls. Interestingly, two of the taxa identified in the significant co-occurrence networks were associated with macronutrients, where fiber intake was associated with higher relative abundances. These findings are aligned with prior observations that SCFA production is determined by the supply of nondigestible carbohydrates.41
Among the limitations of our study is the modest sample size. However, given the expected high inter-individual variability, a thoughtful matching procedure by multiple covariates was performed, which increased the robustness of findings. Nevertheless, the use of MS discordant siblings sharing the same household could have potentially improved confounding control except it may have limited matching for sex and age. Further, despite the minor influence of DMTs observed in the present work, we cannpt rule out its contribution to our findings. Additionally, due to the rarity of POMS, a more conservative threshold for the FDR was used, and significant associations that did not reach multiplicity adjustment threshold were also reported and could represent false-positive findings. Nonetheless, this possibility was minimized by reproducing the study design and analytical procedures in an external cohort. The comparisons between the two datasets could only be made on the genus level since species-level data were annotated as unnamed or uncultured for most of the significant taxa in the complementary set. Additionally, differences in variable collection resulted in slight differences in model adjustment for each dataset limiting result comparability. Last, despite having arisen from a large U.S. nationwide sample of cases and controls, selection bias, a common concerns in case–control studies, cannot be excluded. Moreover, samples were collected close to symptom onset minimizing but not eliminating concerns for reverse causation. Although resource-intensive, population-based approaches with prospective exposure assessment could, in the future, overcome those uncertainties.
Leveraging the unique window of POMS to study disease pathogenesis allowed for the identification of several bacteria that are, either individually or in network clusters, associated with disease status. The contribution of known and predicted SCFA taxa producers had lower abundance in MS cases. Blood and stool measurements of the related metabolites in future studies are warranted to confirm those findings. In summary, our results suggest SCFA producers may be important contributors to MS onset.
Author Contributions
EW conceived the study design and obtained funding. VAS and XZ drafted the first version of the manuscript. XZ conducted, VAS and FZ contributed to the data analysis. SL oversaw the 16S RNA sequencing. JH supervised the data acquisition process. All authors contributed to the data interpretation and manuscript editing, and approved the final version of the manuscript.
Acknowledgments
The authors are grateful for the invaluable contributions of the study subjects and their caregivers. The authors also acknowledge the important contributions of: Michael Sargent (Department of Internal Medicine, and the University of Manitoba IBD Clinical and Research Centre laboratory, Winnipeg, MB, Canada) for managing the stool biobank, and Dr. Jessica D. Forbes (University of Toronto, Toronto, Canada) for assisting with the original grant application. We are also grateful to the investigators and study teams at each site who participated in the Canadian Paediatric Demyelinating Disease Network study, including Dr. Yinshan Zhao, Dr. Morag Graham, Dr. Natalie Knox, Dr. Douglas Arnold, and Christine Bonner.
Funding Information
This study was supported in part by the National Institute of Neurological Disorders and Stroke: (F13NS108668; R01NS071463), the National Multiple Sclerosis Society (HC150906233; RG4861A31), and the Multiple Sclerosis Scientific and Research Foundation (#EGID: 2636; PI:Tremlett). The funding sources were not involved in the study design, the collection, analysis, and interpretation of the data, or in the decision to submit this article for publication.
Conflict of Interest
VAS, AW, ZN, AV, JM, and KEM have no conflicts of interest to report. EW has participated in multicenter clinical trials funded by Genentech, Alexion, and Biogen. She has current support from the NIH, NMSS, DoD, PCORI, CMSC, and Race to Erase MS. She does not receive honorarium from companies. HT has, in the last 5 years, received research support from the Canada Research Chair Program, the National Multiple Sclerosis Society, the Canadian Institutes of Health Research, the Multiple Sclerosis Society of Canada, the Multiple Sclerosis Scientific Research Foundation and the EDMUS Foundation (“Fondation EDMUS contre la sclérose en plaques”). In addition, in the last five years, has had travel expenses or registration fees prepaid or reimbursed to present at CME conferences from the Consortium of MS Centres (2018, 2023), the Canadian Neurological Sciences Federation (2023), National MS Society (2018, 2022), ECTRIMS/ACTRIMS (2017–2023), American Academy of Neurology (2019). Speaker honoraria are either declined or donated to an MS charity or to an unrestricted grant for use by HT's research group. LAB has received funding for research unrelated to this work for Biogen, Alexion and Roche sponsored clinical trials, Shore grant, ROHHAD Fight, Inc, and travel funds from the National MS Society, CDC, and NIH. LAB has also acted as a paid consultant to the national Vaccine Injury Compensation Program and Massachusetts Department of Public Health. CC has received funding for research unrelated to this work from Hoffmann La Roche Ltd. MPG has received funding from Roche/Genentech for clinical trial in POMS. BWG served as a consultant for Biogen, EMD Serono, Novartis, Genentech, Celgene/Bristol Meyers Squibb, Sanofi Genzyme, Bayer, Janssen, Labcorp, and Horizon. She served in speaker bureau for Biogen. Dr. BWG also has received grant/research support from the agencies listed in the previous sentence. She serves in the editorial board for BMJ Neurology, Children, CNS Drugs, MS International and Frontiers Epidemiology. RAM receives research funding from: Canadian Institutes of Health Research, Research Manitoba, Multiple Sclerosis Society of Canada, Multiple Sclerosis Scientific Foundation, Crohn's and Colitis Canada, National Multiple Sclerosis Society, Consortium of MS Centers, the Arthritis Society, US Department of Defense. She is a co-investigator on a study funded in part by Biogen Idec and Roche (no funds to her or her institution). BLB serves as a consultant to Roche, Sanofi, UCB, Biogen, and Genzyme. CNB is supported by the Bingham Chair in Gastroenterology. He receives research funding from: Canadian Institutes of Health Research, Research Manitoba, and Crohn's and Colitis Canada. Dr. Bernstein has served on advisory Boards for AbbVie Canada, Amgen Canada, Bristol Myers Squibb Canada, Eli Lilly Canada, Ferring Canada, JAMP Pharmaceuticals, Pendopharm Canada, Roche Canada, Janssen Canada, Sandoz Canada, Takeda Canada, and Pfizer Canada; Consultant for Takeda; Educational grants from Abbvie Canada, Amgen Canada, Bristol Myers Squibb Canada, Eli Lilly Canada, Pfizer Canada, Takeda Canada, and Janssen Canada. Speaker's panel for Abbvie Canada, Janssen Canada, Pfizer Canada, and Takeda Canada. Received research funding from Abbvie Canada, Amgen Canada, Pfizer Canada, Sandoz Canada, and Takeda Canada. ABO has participated as a speaker in meetings sponsored by and received consulting fees from Accure, Atara Biotherapeutics, Biogen, BMS/Celgene/Receptos, GlaxoSmithKline, Gossamer, Janssen/Actelion, Medimmune, Merck/EMD Serono, Novartis, Roche/Genentech, Sanofi-Genzyme; and has received grant support to the University of Pennsylvania from Biogen Idec, Roche/Genentech, Merck/EMD Serono and Novartis. FZ was funded through research grants held by HT, including The Multiple Sclerosis Scientific and Research Foundation. AIM is funded through the MS Society of Canada endMS Doctoral Studentship (EGID: 3246) and The Multiple Sclerosis Scientific and Research Foundation (PI: Tremlett, EGID: 2636). EAY has received research funding from NMSS, CMSC, CIHR, NIH, OIRM, SCN, CBMH Chase an Idea, SickKids Foundation, Rare Diseases Foundation, MS Scientific Foundation, McLaughlin Centre, Leong Center, Peterson Foundation. Investigator initiated research funding from Biogen. Scientific advisory: Hoffman-LaRoche. Speaker honoraria: Biogen, Saudi Epilepsy Society, NYU, MS-ATL; ACRS, PRIME, and CNPS. GVD receives research funding from Canadian Institutes of Health Research, Multiple Sclerosis Society of Canada, Genome Canada, and the National Science and Engineering Research Council.
Open Research
Data Availability Statement
The data supporting this study's findings are available from the study team upon a reasonable request to the corresponding author.




