Longitudinal Measurements of Glucocerebrosidase activity in Parkinson’s patients
Statistical analyses were performed by Roy Alcalay, Dandi Zheng and Gen Li.
Data used in the preparation of this article were obtained from the Parkinson’s Progression Markers Initiative (PPMI) database (www.ppmi-info.org/data). For up-to-date information on the study, visit www.ppmi-info.org” and the industry partners of the study “PPMI – a public-private partnership – is funded by the Michael J. Fox Foundation for Parkinson’s Research funding partners Abbvie, Allergan, Avid Radiopharmaceuticals, Biogen, BioLegend, Bristol-Myers Squibb, Celgene, Denali, GE Healthcare, Genentech, GlaxoSmithKline, Lilly, Lundbeck, Merck, Meso Scale Discovery, Pfizer, Piramal, Prevail, Roche, Sanofi Genzyme, Servier, Takeda, Teva, UCB, Verily, and Voyager Therapeutics.” (Golub Capital)”
Funding Information
Funding for this study was primarily provided by the Michael J. Fox Foundation. This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging. The authors thank all PPMI participants for their contribution to PD research.
Abstract
Objective
Reduction in glucocerebrosidase (GCase; encoded by GBA) enzymatic activity has been linked to Parkinson’s disease (PD). Here, we correlated GCase activity and PD phenotype in the Parkinson’s Progression Markers Initiative (PPMI) cohort.
Methods
We measured GCase activity in dried blood spots from 1559 samples of participants in the inception PPMI cohort, collected in four annual visits (from baseline visit to Year-3). Participants (PD, n = 392; controls, n = 175) were fully sequenced for GBA variants by means of genome-wide genotyping arrays, whole-exome sequencing, whole-genome sequencing, Sanger sequencing, and RNA-sequencing.
Results
Fifty-two PD participants (13.4%) and 13 (7.4%) controls carried a GBA variant. GBA status was strongly associated with GCase activity. Among noncarriers, GCase activity was similar between PD and controls. Among GBA p.E326K carriers (PD, n = 20; controls, n = 5), activity was significantly lower in PD carriers than control carriers (9.53 µmol/L/h vs. 11.68 µmol/L/h, P = 0.035). Glucocerebrosidase activity was moderately (r = 0.45) associated with white blood cell (WBC) count. Next, we divided the noncarriers with PD to tertiles based on WBC count-corrected enzymatic activity. Members of the lower tertile had higher MDS-Unified Parkinson’s Disease Rating Scale motor score in the “off” medication examination at year-III exam. Longitudinal analyses demonstrated slight reduction of activity in samples collected earlier on in the study, likely because of longer storage time.
Interpretation
GCase activity is associated with GBA genotype, WBC count, and among p.E326K variant carriers, with PD status. Reduced activity may also be associated with worse phenotype but longer follow up is required to confirm this observation.
Introduction
Variants in the glucocerebrosidase gene (GBA) are common genetic risk factors for Parkinson’s disease (PD). GBA encodes the lysosomal enzyme glucocerebrosidase, which hydrolyzes glucosylceramide and glucosylsphingosine to a glucose residue and ceramide and sphingosine, respectively. Several lines of evidence now link reduced glucocerebrosidase activity with PD, even among noncarriers of GBA variants. Reduced glucocerebrosidase enzymatic activity has been linked to increased alpha synuclein aggregation.1 Furthermore, a number of independent autopsy studies have demonstrated reduction in glucocerebrosidase activity in sporadic PD brains compared to control brains.2-5 We and others have also demonstrated that there is a modest reduction in peripheral glucocerebrosidase enzymatic activity (measured in dried blood spots or monocytes) in sporadic PD compared to controls.6, 7
Carrying GBA variants may affect PD phenotype. PD patients with GBA variants have earlier age of onset and a more rapid motor and cognitive progression than noncarriers with PD.8-11 Furthermore, the severity of the GBA mutation (as defined by the Gaucher disease phenotype in homozygous carriers of the mutation; see Table 1 and Table S1 for specific classification) may be associated with age at onset and the rate of PD progression. Onset is earlier and progression is faster in carriers of more severe mutations compared to those with mild mutations.9, 10, 12
| Variant status1 | n | Mean Glucocerebrosidase activity (µmol/L/h) | P-value (compared to noncarriers) | P-value (Parkinson’s vs. controls) | ||||
|---|---|---|---|---|---|---|---|---|
| Total mean and range | Parkinson’s cases (n) | controls (n) | ||||||
| n | Mean (µmol/L/h) | n | Mean (µmol/L/h) | |||||
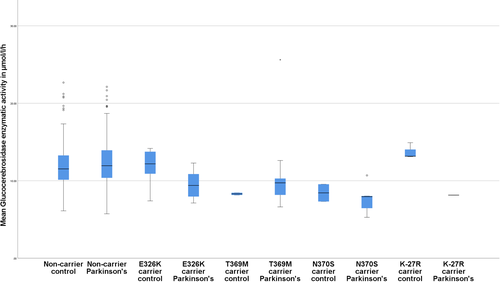
| p.E326K | 25 | 9.96 | 20 | 9.53 | 5 | 11.68 | <0.001 | 0.035 |
| 7.13–14.18 | ||||||||
| p.T369M | 12 | 10.57 | 10 | 11.02 | 2 | 8.30 | 0.0582 | 0.507 |
| 6.63–25.62 | ||||||||
| p.N370S | 9 | 7.75 | 7 | 7.55 | 2 | 8.43 | <0.001 | 0.566 |
| 5.27–10.69 | ||||||||
| p.K-27R | 4 | 12.33 | 1 | 8.15 | 3 | 13.73 | 0.918 | 0.042 |
| 8.15–14.92 | ||||||||
| LRRK2 p. G2019S | 6 | 11.45 | 6 | 12.19 | 0 | n/a | 0.525 | n/a |
| 9.00–16.26 | ||||||||
| Non-GBA non-LRRK2 carriers | 491 | 12.20 | 329 | 12.32 | 162 | 11.95 | n/a | 0.178 |
| 5.73–22.67 | ||||||||
- 1 p.E326K and p.T369M variants are associated with PD, but not with Gaucher’s disease. p.N370S is a mild GBA mutation. p.K-27R is a variant of unknown significance which its association with either PD or Gaucher’s is not established.
- 2 P-value of the differences in WBC count-corrected activity is P < 0.001.
Given the link between GBA variants and disease severity, we tested the association between glucocerebrosidase enzymatic activity and PD phenotype in a cross-sectional study.6 We reported that reduced activity was associated with shorter disease duration (<6 years in lower glucocerebrosidase enzymatic activity tertile vs. 9 years in the higher tertile), suggesting a more rapid progression. To test if reduced glucocerebrosidase enzymatic activity is associated with faster progressing PD phenotype, we proposed analysis of a longitudinal cohort. Here, we present results of a longitudinal study, in which we measured glucocerebrosidase enzymatic activity in dried blood spots in a cohort of individuals with early PD and a non-PD comparator group enrolled in the Parkinson’s Progression Marker Initiative (PPMI) study. We aimed to: (1) test the association between glucocerebrosidase activity, age, and sex and confirm our prior observation of reduced glucocerebrosidase activity in PD; (2) test enzymatic activity in different GBA variants (3) explore the utility of longitudinal measurements of glucocerebrosidase activity as a biomarker, and the effect of time (aging) on the activity; and (4) test if reduced glucocerebrosidase activity predicts rates of motor and cognitive progression.
Material, Methods and Participants
Participants and clinical evaluation
PPMI is an international, multiple-site, prospective, longitudinal cohort study. Participants in this study are from the inception PPMI cohort who had frozen whole blood available from which glucocerebrosidase enzymatic activity could be measured. In brief, newly diagnosed, de-novo (untreated) Parkinson’s patients (N = 423) and generally healthy controls (N = 196) were enrolled from June 2010—May 2013. At baseline, participants with PD were required to have a dopamine transporter (DAT) deficit on imaging. The aims and methodology of the study have been published elsewhere13 and are available at www.ppmi-info.org/study-design. The overall study was approved by the Research Subjects Review Board at the University of Rochester, and the study was approved by the institutional review board at each site. Participants provided written informed consent at each visit.
Glucocerebrosidase enzymatic activity assay
Frozen whole blood received from the PPMI biospecimen repository was thawed on watered ice gently rocking for 45–60 min. In sum, 1559 samples collected at four annual visits (baseline and years 1–3) from 567 participants (including 392 PD participants and 175 controls) were available for analysis. All samples were shipped to a central repository and stored in a −80°C freezer. Once thawed, the blood was spotted as previously reported for dried blood spots (DBS) prepared from fresh blood.14, 15 In brief, 75 μL of the blood was spotted on Whatman 903™ filter paper (GE Healthcare, Piscataway, NJ) dropwise, then dried overnight in a well ventilated area. Dried blood spots prepared from frozen blood (FB DBS) were sealed in plastic bags with desiccant (Uline, Pleasant Prairie, WI) and humidity indicator cards (Static Control, Sanford, NC) and stored at −80°C until analysis.
Glucocerebrosidase enzymatic activity was measured using a previously published protocol as part of a multiplex assay with four additional lysosomal enzymes.16 In summary, glucocerebrosidase was extracted from a 3.2 mm-diameter punch per FB DBS sample in 70 µL of 20 mmol/L sodium phosphate buffer (pH 7.1) on a 96-well plate. Ten µL of the FB DBS extract was added to 15 µL of glucocerebrosidase substrate/internal standard mixtures (0.67 mmol/L of C12-glucocerebroside and 13.33 μmol/L C14-ceramide, Center for Disease Control and Prevention, Georgia, Atlanta) in citrate-phosphate buffer (0.31/0.62 mol/L, Sigma, St. Louis, MI) with sodium taurocholate (16 g/L, Sigma St. Louis, MI). Sealed plates were incubated on an orbital shaker at 37°C for 20–24 h. Reactions were quenched with 100 µL of organic solution (1:1 ethyl acetate:methanol) and followed with liquid–liquid and solid phase extractions. The samples were dried under nitrogen, sealed and stored at −80°C. Prior to tandem mass spectrometry (MS/MS) analysis, plates were thawed and reconstituted with 200 µL of a solvent mixture (80:20 acetonitrile:water containing 0.2% formic acid).
All analytes were monitored on an API 5000 triple quadrupole mass spectrometer (ABSciex, Framingham, Massachusetts, USA) by Multiple Reaction Monitoring (MRM). The enzyme activity of each sample was calculated from the ion abundance ratio of product to internal standard as measured by the mass spectrometer. Background activity of a blank filter paper was subtracted from the DBS or FB DBS activity. Activity was expressed as micromoles of product per liter of whole blood per hour (µmol/L/h). Two DBS samples with previously established normal activity levels for each enzyme, as well as DBS samples with 5% residual enzyme activity representative of disease controls (Newborn Screening Quality Assurance Program, Center for Disease Control and Prevention, B1508) were included in each plate for quality control (QC). All scientists were blinded to all clinical data including Parkinson’s disease and genetic status.
Sequencing and genotyping of GBA variants and LRRK2 G2019S
Participants recruited into the PPMI de novo cohort and healthy controls did not have genotyping at the time of recruitment, but had it performed later in the study by the Genetic Core. Genotyping data were obtained from a number of assays using DNA or RNA. Assays included: genome-wide genotyping arrays (GWAS), whole-exome sequencing (WES), whole-genome sequencing (WGS), Sanger sequencing, and RNA-sequencing (RNA-seq). Genotype data were processed to extract LRRK2 p.G2019S and coding variants for GBA. For a table of included variants and which genetic assays covered each variant, see Table S2. Genotype data were obtained as described in the following section. More information about these assays has also been previously published.17 Numbers of participants with each data type are listed in Table S3; the majority of participants analyzed were covered on all five platforms (458 out of 562, excluding five participants below), with small numbers of participants missing one or more platforms. One participant was reported to carry the GBA p.L444P variant which was not resolved following manual review; this participant was excluded from the genetic analyses. Four additional participants had missing data for LRRK2 and/or GBA variants and were also excluded.
GWAS (Project 107; accessed 11/2013), Sanger (Project 126; accessed 07/2019), and WES (Project 116; accessed 02/2015) data were downloaded from the Laboratory of NeuroImaging (LONI) Image Data Archive PPMI project website (https://ida.loni.usc.edu/). WGS data were obtained from Dr. Andrew Singleton’s laboratory at the National Institutes of Health’s National Institute on Aging, which is also posted on LONI (Project 118; July2018 version). RNA-seq data were obtained from Dr. David Craig’s laboratory at the University of Southern California Keck School of Medicine; however, these data are also posted on LONI (Project 133). Detailed information on these genetic data are available as methods documents from LONI.
Briefly, GWAS assays were performed using the Illumina Infinium iSelect HD custom genotyping NeuroX array18 (Illumina, Inc., San Diego, CA; 267,607 standard exonic variants and 24,706 custom variants). Five hundred and nineteen of the 562 participants included in these analyses have GWAS data. WES was performed on 534 of the 562 participants using the Illumina Nextera Rapid Capture Expanded Exome kit and the Illumina HiSeq 2500 sequencing platform. WGS for paired-end reads has been performed on the majority of PPMI participants (N = 558) using the Illumina TruSeq PCR Free DNA Sample Preparation Guide and the Illumina HiSeq X Ten Sequencer. Sanger sequencing was performed to capture GBA exons 1–11, with data for nine variants available on LONI (for list see Table S2). Whole-transcriptome RNA-seq was performed by Hudson Alpha’s Genomic Services Laboratory on ribosomal and globin-depleted blood derived RNA collected at multiple visits using PaxGene tubes from PPMI participants. Samples were sequenced on the Illumina NovaSeq6000; processed count data are available on LONI, with raw data available on request.
All variant data were compiled into a database. For variant calls that differed between genetic assays, originating data were reviewed where needed and possible (i.e., for WGS, WES, and RNA-seq, read depth and allele counts were reviewed and data were viewed in the Integrative Genomics Viewer (IGV; https://software.broadinstitute.org/software/igv/19-21), and manual curation was completed to create a consensus variant call for the participant. RNA-seq was not used as a primary source of variant data, due to concerns about shallow read depth in some samples and variants as well as concern about transcribed allele bias. However, RNA-seq was used as a secondary data source to resolve several discrepancies in other genetic platforms; the longitudinal samples collected for most participants was a further data source to confirm genotype where necessary.
Statistical analysis
Glucocerebrosidase enzymatic activity and age, sex, WBC count, and Parkinson’s status
We predicted that glucocerebrosidase activity is associated with white blood cell (WBC) count, because the activity is measured in leukocytes. WBC count is available from each PPMI site laboratory for each study visit. We therefore correlated glucocerebrosidase activity and WBC count. Since they were moderately correlated (r = 0.45), we computed a WBC-corrected variable: glucocerebrosidase activity at a study visit/ WBC count at the same study visit. Glucocerebrosidase activity was available from four annual visits (baseline to year 3). To assign each participant a single glucocerebrosidase activity value, we averaged glucocerebrosidase activity per participant and computed a mean glucocerebrosidase activity variable. We did not use the baseline measurement only because it was missing for 170 participants. Similarly, we averaged the WBC count-corrected glucocerebrosidase activity per participant.
We then tested the association between glucocerebrosidase activity and WBC-count corrected glucocerebrosidase activity with (1) demographics, (2) PD status, and (3) GBA variant status.
GBA variants and glucocerebrosidase enzymatic activity
We present data on glucocerebrosidase activity on each variant separately. For the p.E326K variant, there were sufficient cases and controls to compare glucocerebrosidase activity between carriers with and without PD.
Longitudinal measurements of glucocerebrosidase enzymatic activity
To demonstrate the longitudinal measurements of glucocerebrosidase activity per participant, we plotted the activity over time in spaghetti plots. To test the association between longitudinal measures of glucocerebrosidase enzymatic activity (and enzymatic activity corrected for WBC count) and covariates, a linear mixed-effect model (LMM) with random intercept was employed. The covariates in the model include sex, PD status (PD vs. control), time (PPMI study visit), age at baseline, GBA variant status (binary yes/no), and the interaction term between diagnosis status and time. Analyses were first conducted on all participants. Then, GBA carriers, LRRK2 carriers and participants whose PD diagnosis changed over the study course (e.g., from PD to Progressive Supranuclear Palsy, PSP) were excluded for the next analyses.
Glucocerebrosidase enzymatic activity and Parkinson’s phenotype in noncarriers
To test whether changes in glucocerebrosidase activity predict the progression of motor and cognitive change over time in sporadic PD, LMMs with random intercept were implemented only in PD participants without LRRK2 or GBA variants. To test cognitive changes over time, the longitudinal MoCA score (outcome) was tested in models including sex, time, age at baseline, education in years, ApoE status, and each of the following predictors: (i) Glucocerebrosidase enzymatic activity; (ii) WBC-count corrected glucocerebrosidase activity; (iii) WBC count. We also used LMMs with random intercept to associate the Movement Disorders Society (MDS) Unified Parkinson's Disease Rating Scale (UPDRS) Part II and the MDS-UPDRS Part III with the above predictors in PD participants, adjusting for sex, age at baseline, and levodopa equivalent daily dose.
Next, we divided the PD participants without LRRK2 or GBA variants into tertiles based on mean glucocerebrosidase activity corrected for WBC count, and compared demographics and clinical characteristics among tertiles. We chose a division to tertiles to be able to compare to our previously reported cross-sectional analysis.6 For motor phenotyping, we used the MDS-UPDRS, Part II and Part III. For cognitive phenotyping, we used the MoCA, and two previously computed categorical variables: The first divided the cohorts into “impaired” and “not-impaired” based on neuropsychological performance, as previously reported by Caspell-Garcia and colleagues;22 the second categorical variable was based on a clinical assessment of the participants as “normal,” “mild cognitive impairment” or “dementia.” For the purpose of the analyses, we dichotomized the analysis to “normal” versus “mild cognitive impairment or dementia”.
Lastly, logistic regression was used to associate neuropsychological performance in year 4 or cognitive impairment on clinical assessment in year 3 (clinical data on year 4 was not complete) with glucocerebrosidase activity in PD patients. Binary outcome was regressed on sex, age at baseline, educations in years, ApoE status and the following predictors: (i) the average glucocerebrosidase activity; and (ii) the average glucocerebrosidase activity over WBC count. We repeated the analyses with and without GBA and LRRK2 variant carriers to test if GBA mutation status was associated with worse cognitive outcome.
Of note, we have not applied statistical tools to test for multiple comparisons. Analyses were performed using R Statistics version 3.5.1 software, SPSS version 24 software and Statistical Analysis System (SAS) statistical software package, version 9.4, SAS institute Inc.
Results
Glucocerebrosidase enzymatic activity and age, sex, WBC count, and Parkinson’s status
Glucocerebrosidase activity was moderately associated with WBC count measured from the same sample collections (e.g., baseline visit: r = 0.44, P < 0.001). The association between glucocerebrosidase activity and WBC count (including differential of neutrophil, monocyte, lymphocyte, basophil, and eosinophil counts) is described in Table S4. Because of this association, further analyses were conducted using crude values and WBC count-corrected values. Female sex was associated with both lower WBC count and mean glucocerebrosidase activity, but WBC count-corrected activity was similar between the sexes (males: 2.07 µmol/L/h corrected for WBC count, SD = 0.48; females: 2.06 µmol/L/h corrected for WBC count, SD = 0.55; P = 0.877). There was no association between glucocerebrosidase activity and participant age (r = −0.043; P = 0.368).
The demographics, genotype, and glucocerebrosidase activity of the study participants are included in Table 2. Of note, there were seven LRRK2 p.G2019S mutation carriers (1.8%) and 52 (13.4%) GBA variant carriers among the Parkinson’s participants versus no LRRK2 mutation carriers and 13 (7.4%) GBA-variant carriers in controls.
| Parkinson’s cases (n = 392) | Controls (n = 175) | P-value | |
|---|---|---|---|
| Mean age in years at recruitment, (SD) | 61.8 (9.5) | 60.9 (11.4) | 0.304 |
| Males % (n) | 65.3% (256) | 66.9% (117) | 0.774 |
| LRRK2 p.G2019S carriers, % (n)1 | 1.8% (7) | 0.0% (0) | 0.105 |
| GBA variant carriers, % (n)2 | 13.4% (52) | 7.4% (13) | 0.046 |
| Mean Glucocerebrosidase enzymatic activity in µmol/L/h (SD) | 11.89 (3.11) | 11.91 (2.93) | 0.951 |
| Mean Glucocerebrosidase enzymatic activity corrected to white blood count in µmol/L/h corrected for WBC count (SD) | 2.05 (0.52) | 2.12 (0.46) | 0.139 |
- 1 LRRK2 p.G2019S mutation status is missing on four Parkinson’s cases.
- 2 GBA variant status is missing on three Parkinson’s cases (two of which are also missing LRRK2 p.G2019S status).
GBA variants and glucocerebrosidase enzymatic activity
The different genotypes and the associated mean glucocerebrosidase activity are described in Table 1 and Figure 1 for variants present in three or more participants and in Supplementary Table 1 for genotype present in 1–2 participants.
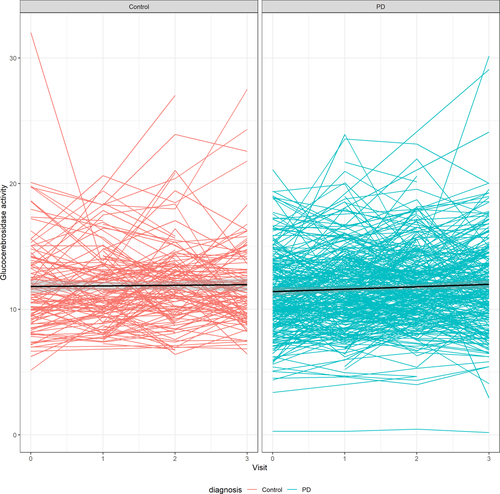
Longitudinal measurements of glucocerebrosidase enzymatic activity
The change in glucocerebrosidase enzymatic activity over time (raw and corrected for WBC count) is presented in Figures 2, 3. In the linear mixed effect model with glucocerebrosidase enzymatic activity as an outcome, activity was significantly associated with sex (estimate −0.597; SD = 0.257; P = 0.0203) and GBA genotype (estimate −2.830; SD = 0.383; P < 0.0001), but not with PD diagnosis (P = 0.955), visit (P = 0.128), age at enrollment (P = 0.109) or the interaction term between PD diagnosis and visit (P = 0.456). Visit number was significantly associated with glucocerebrosidase activity among PD patients (estimate 0.240; SD = 0.06; P = 0.0001), but not in the control group (estimate 0.155; SD = 0.105; P = 0.1422) when analysis was stratified by PD status.
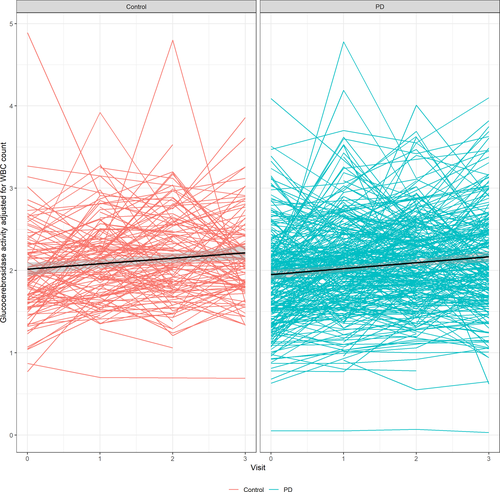
In the linear mixed effect model with WBC count-corrected glucocerebrosidase activity as an outcome, WBC count-corrected glucocerebrosidase activity was significantly associated with visit number (estimate 0.063; SD = 0.020; P = 0.0008) and GBA genotype (estimate −0.600; SD = 0.062; P < 0.0001), but not with sex (P = 0.426), PD diagnosis (P = 0.454), age at enrollment (P = 0.144) or the interaction term between PD diagnosis and visit (P = 0.548). When analysis was stratified by PD status, visit number was associated with WBC count-corrected enzymatic activity in both populations (PD: estimate 0.077; SD = 0.012; P < 0.0001; Control: estimate 0.065; SD = 0.019; P = 0.0007). Based on these values, assuming increased activity over time is a result of a shorter “freezer time” in newer samples, for every year in the freezer, we estimate a 0.151 µmol/L/h decrease in glucocerebrosidase enzymatic activity and 0.063 µmol/L/h corrected to WBC count annual decrease in glucocerebrosidase activity corrected to WBC count, adjusting for other variables.
Glucocerebrosidase enzymatic activity and Parkinson’s phenotype in non-carriers
In linear mixed effect models, glucocerebrosidase activity was not associated with MDS-UPDRS “on” (P = 0.711) or “off” (P = 0.945) examinations or with MoCA scores (P = 0.628) in adjusted models. Glucocerebrosidase enzymatic activity corrected to WBC count was also not associated with MDS-UPDRS (P = 0.438), but was inversely associated with MoCA scores, when MoCA scores were the outcome (estimate −0.477, SD = 0.157, P = 0.0026), in a model adjusted for sex, visit, age, education and ApoE status. Visit (P = 0.0028), age at enrollment (P < 0.0001), education (0.0437), but not sex (P = 0.552) or ApoE4 status (P = 0.479), were associated with MoCA performance. This would suggest that incremental increases in WBC count-corrected glucocerebrosidase enzymatic activity over time are associated with reduced MoCA performance.
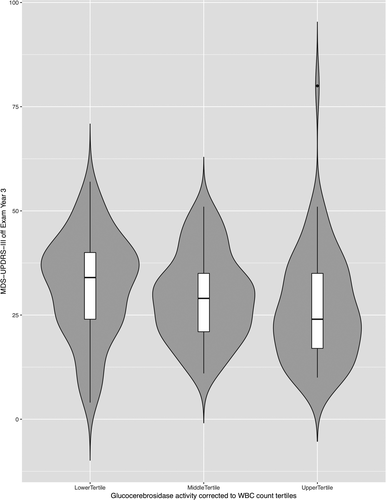
Next, we divided the non-LRRK2 non-GBA PD cohort to tertiles based on mean glucocerebrosidase activity corrected to WBC count, and compared the motor and cognitive phenotypes in year 3 visit among the tertiles (Table 3). Of note, there was no demographic differences among the tertiles, but MDS-UPDRS-III scores in the “off” exam were significantly higher in the lower tertile group compared to the upper tertile group (Figure 4). A similar trend, albeit not significant, was noted in the MDS-UPDRS-III “on” exam and the MDS-UPDRS total (II + III) exam in the off state. There was no difference across the tertiles in MoCA score and the categorical variables of cognition (neuropsychological performance and cognitive impairment on clinical assessment).
| Lower GCase tertile (n = 108) | Middle GCase tertile (n = 109) | Higher GCase tertile (n = 108) | P-value | |
|---|---|---|---|---|
| Mean GCase activity in µmol/L/h corrected to WBC count, (SD) | 1.62 (0.21) | 2.11 (0.11) | 2.65 (0.26) | <0.001 |
| Range | 0.70–1.88 | 1.88–2.30 | 2.30–3.41 | |
| Mean GCase activity in µmol/L/h, (SD) | 11.07 (2.41) | 12.18 (2.50) | 13.64 (2.87) | <0.001 |
| Range | 5.73–18.39 | 7.61–20.93 | 8.76–22.13 | |
| Percent male, (n) | 65.7% (71) | 71.6% (78) | 64.8% (70) | 0.516 |
| Mean age at recruitment in years, (SD) | 63.9 (9.9) | 62.5 (8.8) | 61.0 (9.2) | 0.073 |
| Education in years, (SD) | 15.6 (3.2) | 15.8 (2.9) | 15.4 (2.6) | 0.625 |
| Levodopa equivalent daily dose in mg at Year 3, (SD)1 | 447 (455) | 416 (262) | 467 (336) | 0.642 |
| MDS-UPDRS-III off exam Year 3, (SD)2 | 32.0 (12.1) | 29.3 (10.2) | 26.6 (13.0) | 0.0352 |
| MDS-UPDRS-III on exam Year 3, (SD) 3 | 26.4 (12.2) | 24.3 (11.0) | 22.6 (12.9) | 0.106 |
| MDS-UPDRS total off exam Year 3, (SD) 2 | 50.1 (19.0) | 46.5 (15.6) | 42.8 (20.1) | 0.075 |
| MoCA Year 3, (SD)4 | 26.2 (3.2) | 26.8 (2.7) | 26.7 (2.5) | 0.335 |
| Percent impaired in CogState Year 3 (n)5 | 25.3% (23) | 22.9% (22) | 20.0% (18) | 0.698 |
| Percent impaired by neuropsychological testing in Year 3 (n)6 | 16.1% (15) | 21.1% (20) | 13.2% (12) | 0.349 |
- 1 Available on 265 participants.
- 2 Available on 200 participants; Differences between the lower and upper tertile drive the significance based on Bonferroni post hoc analysis.
- 3 Available on 264 participants.
- 4 Available on 200 participants.
- 5 Available on 214 participants.
- 6 Available on 232 participants.
Lastly, we applied logistic regression to test if glucocerebrosidase activity or glucocerebrosidase activity adjusted to WBC count are associated with cognitive impairment in models adjusted for sex, age at baseline, educations in years, and ApoE status (yes/no carrier of ApoE4). In the models where the outcome was cognitive impairment by clinical assessment (which was dichotomized to “normal” vs. “mild cognitive impairment” or “dementia”) in year 3, age (P < 0.001), but no other covariate was associated with mild cognitive impairment or dementia, including mean glucocerebrosidase activity (unadjusted to WBC count, P = 0.516; adjusted to WBC count, P = 0.894). In the models where the outcome was neuropsychological performance in year 4 (i.e., impaired vs. not impaired, based on computation previously reported by Caspell-Garcia and colleagues22), age (P = 0.006), education (P = 0.020) and ApoE status (P = 0.046) were associated with impaired performance, but not glucocerebrosidase activity (unadjusted to WBC count, P = 0.323; adjusted to WBC count, P = 0.502). Lastly, when the models included all PD participants, GBA variant status was also not associated with membership in the impaired group based on clinical diagnosis in year 3 (P = 0.138) or with impaired neuropsychological performance in year 4 (P = 0.125).
Discussion
This is the first report of repeated measurements of glucocerebrosidase enzymatic activity in a deeply phenotyped PD cohort. Our findings can be divided to those which replicate our findings from a cross-sectional study, findings that we were not able to replicate, and novel findings given the longitudinal nature and wealth of data availability on the PPMI cohort. Similar to our findings in a cross-sectional report, glucocerebrosidase activity is strongly correlated with GBA genotype, including the more common non-Gaucher disease causing variants linked to PD, p.E326K and p.T369M. As before, we did not find an association between sex, age, and glucocerebrosidase activity. Furthermore, there is no evidence that glucocerebrosidase activity decreases with time (age). This study failed to replicate our previous finding of reduced glucocerebrosidase activity in noncarriers with PD compared to noncarrier controls; this may be a result of the current sample size, which is smaller than the cross-sectional study thus diminishing the statistical power.6 Alternatively, it is possible that the differences in disease duration between the two cohorts (longer duration in the 2015 study) may explain the discrepant findings. In contrast, among p.E326K carriers, activity was even further reduced in carriers with Parkinson’s (9.53 µmol/L/h) compared to p.E326K carriers without PD (11.68 µmol/L/h). This finding should be reviewed with caution, since this analysis is based on small n of p.E326K carriers (n = 25). The difference between glucocerebrosidase activity in p.E326K carriers with PD and carriers without PD (roughly 20%) are higher than the differences we previously reported in activity between idiopathic PD carriers and noncarriers controls (roughly 6%). If these differences between GBA variant carriers with PD compared to variant carriers without PD are replicated in larger cohorts of variants carriers, it would imply that alterations of glucocerebrosidase activity may be more meaningful among variant carriers than among noncarriers.
Our study demonstrated that enzymatic activity measured in dried blood spot is moderately correlated with WBC count (r = roughly 0.45). Therefore, we recommend adjusting for WBC count in future studies of peripheral glucocerebrosidase activity as a biomarker for PD. When different cell types were correlated individually, both neutrophil and monocyte counts were associated with glucocerebrosidase activity, but not lymphocytes, eosinophils, or basophils were not (Table S4). Lastly, PD patients who are noncarriers with lower mean enzymatic activity performed worse on “off” motor examination during year 3 evaluation, supporting an association of glucocerebrosidase activity with disease severity in non-GBA non-LRRK2 variant carriers, but given the multiple analyses conducted, this finding needs to be confirmed with a longer follow-up.
Glucocerebrosidase enzymatic activity by GBA variant status
This study demonstrates the high burden of glucocerebrosidase alterations on PD. In this cohort of early, untreated PD, 13.4% of PD participants carry a GBA variant. GBA genotyping coupled with glucocerebrosidase activity measurements may help clarify the pathogenicity of certain variants (see Table S1), and confirms that common variants in GBA that are associated with PD are associated with reduced activity compared to noncarriers (p.E326K; p.T369M).
The glucocerebrosidase enzymatic activity for the single GBA p.N370S homozygote analyzed clearly stood apart from heterozygotes and noncarriers (Figures 2, 3), and similar to our previous report, glucocerebrosidase activity differed significantly between carriers and noncarriers. While GBA variant carriers have significantly lower activity than noncarriers, there is a subset of participants without GBA variants whose activity overlaps with heterozygotes. We hypothesize that additional genes and possibly environmental factors are associated with the reduced activity, and these should be investigated in follow-up studies. Studies including large numbers of nonmanifesting carriers of GBA variants (e.g., p.N370S) are required. Such studies would allow the comparison of enzymatic activity in heterozygote carriers of different variants with and without PD. If these studies confirm our finding of reduced activity in variant carriers with PD compared to variant carriers without PD, this would suggest that glucocerebrosidase activity may partially explain the incomplete penetrance of PD among GBA carriers, and that lower activity is associated with a greater risk.
Longitudinal measurements of glucocerebrosidase enzymatic activity
Repeated measurements of glucocerebrosidase activity in frozen blood dried blood spots demonstrated substantial variance, and an overall small but significant trend toward lower activity with longer freezer time. Therefore, while frozen sample dried blood spot may be a crude biomarker of glucocerebrosidase reduction in PD, more sensitive methodologies (e.g., as proposed by Atashrazm and colleagues7) may be required to identify subtle differences between PD patients and controls without GBA variants. Correlating longitudinal changes in glucocerebrosidase activity with longitudinal changes in motor and cognitive functioning failed to demonstrate an association. This finding may be explained by the relative stability of glucocerebrosidase activity over time, in contrast to progression of motor and cognitive deficits in PD.
Glucocerebrosidase enzymatic activity and clinical phenotype in sporadic PD
We have not found a strong correlation between glucocerebrosidase enzymatic activity and PD phenotype. Surprisingly, in the longitudinal models, increased activity was inversely associated with MoCA performance, but a similar association was not observed with neuropsychological performance or clinician assessment of cognitive impairment. It is possible that longer disease duration is required to distinguish phenotypic dissociation between those with low versus high enzymatic activity. Similarly, using the same outcomes, GBA variant status was not associated with worse cognitive performance after 3–4 years follow-up, even though GBA genotype is clearly associated with worse cognitive functioning as PD progresses.23 In contrast, motor scores on the MDS-UPDRS Part III “off” were higher (worse) among those in the lower tertile of activity compared to those in the higher tertile of glucocerebrosidase enzymatic activity (32.0 vs. 26.6, P = 0.035). Similar, albeit insignificant, trends were observed in MDS-UPDRS questionnaire (UPDRS-II) and the total scores. This finding suggest that glucocerebrosidase activity may be associated with motor outcome, but given the multiple comparisons conducted, longer follow-up is required to confirm these findings.
Our results may inform the design of future clinical trials. The discovery of the link between GBA variants and PD highlights the potential role of the lysosome in the pathophysiology of PD, and provides a drug target for disease-modifying interventions. Multiple clinical trials targeting the glucocerebrosidase pathway in PD are ongoing world-wide (e.g., ClinicalTrials.gov Identifier: NCT02914366, NCT02941822, and NCT02906020;24, 25). Assuming such interventions may be able to modify disease progression, it remains unknown who would benefit from them. Currently, trials are focused on GBA variant carriers, but given relative reduction of glucocerebrosidase in sporadic PD brains,2-5 it is conceivable that noncarriers may also benefit from glucocerebrosidase-targeted interventions. Linking reduced glucocerebrosidase with PD phenotype (i.e., faster progression like in GBA carriers) may be useful in future patient selection (or stratification) in clinical trials. The comprehensive longitudinal design of the PPMI study makes it ideal for biomarker testing.
Strengths, limitations, and future studies
The strengths of our study are the relatively large and very carefully phenotyped longitudinal cohort analyzed, and the blinding of the laboratories performing enzymatic analyses. Also, the data are publicly available (Table S5 and LONI). In addition, the study replicated previous observations (as above), which reinforce the results. Limitations include the sample size, especially of GBA variant carriers without PD, and the multiple comparisons conducted.
Summary
In summary, glucocerebrosidase enzymatic activity correlates with GBA variant status and WBC count, which should be adjusted for in future PD biomarker studies. We identified an association between reduced activity and worse phenotype among noncarriers of GBA variants, but a longer follow up is required to confirm if low dried blood spot activity predicts accelerated motor progression. In accordance with the Michael J. Fox Foundation policy, enzymatic activity, as well as all other datapoints, are publically available for future analysis.
Acknowledgments
Funding for this study was primarily provided by the Michael J. Fox Foundation. This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging. The authors thank all PPMI participants for their contribution to PD research.
Conflicts of Interest
Pavlina Wolf, Kate Zhang, Ming Sum, Ruby Chiang, Karolina Helesicova and Sergio Pablo Sardi are/were employees of Sanofi during their work on the manuscript. All other authors have no conflict of interest related to this work.
Disclosures
Roy Alcalay is funded by the NIH, the Parkinson’s Foundation and the Michael. J. Fox Foundation. He received consultation fees from Sanofi, Roche, Janssen and Restorbio. Dr Gan-Or is supported by the Fonds de recherche du Québec - Santé (FRQS) Chercheurs-boursiers award given in collaboration with Parkinson Quebec, and is a Parkinson Canada New Investigator awardee. He received consultancy fees from Lysosomal Therapeutics Inc. (LTI), Idorsia, Prevail Therapeutics, Inceptions Sciences (now Ventus), Denali, Ono Therapeutics and Deerfield. Tanya Simuni, MD has served as a consultant for Acadia, Abbvie, Accorda, Adamas, Allergan, Amneal, Anavex, Aptinyx, Blue Rock Therapeutics, Denali, General Electric (GE), Neuroderm, Neurocrine, Sanofi, Sinopia, Sunovion, TEVA, Takeda, Voyager, US World Meds, Parkinson’s Foundation, and the Michael J. Fox Foundation for Parkinson’s Research; Dr. Simuni has served as a speaker and received an honorarium from Acadia and Adamas; Dr Simuni is on the Scientific advisory board for Anavex, Neuroderm, Sanofi, and MJFF. Dr. Simuni has received research funding from the NINDS, Parkinson’s Foundation, MJFF, Biogen, Roche, Neuroderm, Sanofi, Sun Pharma, Abbvie, and IMPAX. Lana M. Chahine receives research support from the Michael J Fox Foundation, including for PPMI, has received travel payment from MJFF to MJFF conferences, is a paid consultant to MJFF, receives research support from the UPMC Competitive Medical Research Fund, is study site investigator for a study sponsored by Biogen, is a subinvestigator for a study sponsored by Voyager, and receives royalties from Wolters Kluwel (for book authorship). Cornelis Blauwendraat is an NIH employee and reports no disclosures.
Parkinson’s Progression Marker Initiative Authors
Steering Committee
Kenneth Marek, MD1 (Principal Investigator); Andrew Siderowf, MD MSCE12 (co-PI); Danna Jennings, MD53 (Industry Scientific Advisory Board); Caroline Tanner, MD, PhD9 (Site Investigator); Tanya Simuni, MD3 (Site Investigator); Christopher Coffey, PhD4 (Statistics Core, PI); Karl Kieburtz, MD, MPH5 (Clinical Core, PI); Werner Poewe, MD7 (Site Investigator); Brit Mollenhauer, MD8 (Bioanalytics Core, co-PI; Site Investigator); Douglas Galasko, MD27 (Bioanalytics Core, co-PI; Site Investigator); Tatiana Foroud, PhD15 (Genetics Coordination Core and Biorepository Core, PI); Todd Sherer, PhD6; Sohini Chowdhury6; Mark Frasier, PhD6; Vanessa Arnedo6; Jamie Eberling, PhD6; Samantha J. Hutten, PhD6; Alyssa N. Reimer6; Luba Smolensky, MS6; Dave Alonso6; Chris Baglieri, MS66 (Advanced Analytics Core, PI); Amanda Christini, MD66; Susan Bressman, MD14 (Site Investigator); Ray Dorsey, MD MBA5 (Clinical Core); Cynthia Casaceli, MBA5 (Clinical Core); Paola Casalin11 (Biorepository Core); Giulia Malferrari11 (Biorepository Core); Lana Chahine, MD67; Nichole Daegele, MA1 (Leadership Core); Margaret Bockus, MA5 (Clinical Core); Raymond James, RN22 (Site Coordinator); Sugi Mahes5 (Clinical Core); Kalpana Merchant, PhD68; Kathleen Poston, MD65 (Imaging Core); Ekemini Riley, PhD69; John Seibyl, MD1 (Imaging Core, PI); Leslie Shaw, PhD12; Andrew Singleton, PhD13 (Genetics Core, PI); Arthur Toga, PhD10 (Bioinformatics Core, PI); Duygu Tosun-Turgut, PhD9 (DTI Analysis Core, PI); John Trojanowski, MD PhD12; Dan Weintraub, MD12, Emily Flagg1 (Leadership Core), Roseanne Dobkin, PhD70, Ethan Brown, MD9.
Study Cores
Leadership Core: Kenneth Marek, MD1; Nichole Daegele1.
Clinical Coordination Core: Cynthia Casaceli, MBA17; Ray Dorsey, MD, MBA17; Renee Wilson17; Sugi Mahes17.
Imaging Core: John Seibyl, MD1; Christina Salerno1.
Statistics Core: Chelsea Caspell4; Liz Uribe4; Janel Fedler, PhD4; Michael Brumm, MS4; David Erick LaFontant, MS4.
Bioinformatics Core: Karen Crawford10.
Biorepository: Mali Gana Weisz, PhD61; Avi Orr-Urtreger, MD PhD61.
Bioanalytics Core: John Trojanowski, MD, PhD2; Leslie Shaw, PhD2.
Genetics Core: Andrew Singleton, PhD7.
Genetics Coordination Core: Tatiana Foroud, PhD13.
Pathology Core: Tatiana Foroud, PhD13; Thomas Montine, MD, PhD14.
Wearables Core: Tatiana Foroud, PhD13.
Advanced Analytics Core: Chris Baglieri65, Amanda Christini, MD65.
Site Management Core: Lisa de Blieck, MPA1, Whitney Richardson, MS1.
Site Investigators
David Russell, MD, PhD1; Stewart Factor, DO16; Penelope Hogarth, MD17; David Standaert, MD, PhD18; Robert Hauser, MD, MBA19; Joseph Jankovic, MD20; Nabila Dahodwala, MD12; Marie H Saint-Hilaire, MD22; Irene Richard, MD23; Klaus Seppi, MD7; David Shprecher, MD24; Hubert Fernandez, MD25; Kathrin Brockmann, MD26; Isabel Wurster MD26; David Brooks, MD29; Yen Tai29; Paolo Barone, MD, PhD30; Stuart Isaacson, MD31; Alberto Espay, MD, MSc32; Dominic Rowe, MD, PhD33; Christopher Way, DO2; Eduardo Tolosa MD34; Shu-Ching Hu, MD, PhD21; Douglas Galasko MD27; Emile Moukheiber28; Jean-Christophe Corvol, MD39; Nir Giladi, MD61; Javier Ruiz Martinez, MD60; Jan O. Aasly, MD62; Leonidas Stefanis, MD PhD63; Karen Marder, MD MPH64.
Coordinators
Julie Festa1; Cheryl Riordan1; Katrina Wakeman; Alison Freed17; Karen Williams3; Courtney Blair18; Leigh Harrell19; Christine Hunter, RN20; Krista Specketer21; Cathi-Ann Thomas, RN, MS22; Nicole Guerrero23; Beatrice Heim, MD7; Diana Willeke8; Victoria Brown24; Jennifer Mule25; Ella Hilt26; Shawnees Peacock, MS27; Arita McCoy28; Kori Ribb28; Susan Ainscough30; Lisbeth Pennente31; Christina Gruenwald32; Madelaine Ranola33; Barbara Sommerfeld MSN RN16; Farah Kausar9; Alicia Garrido, MD34; Deborah Raymond14; Benjamin Le Toullec39; Anat Mirelman, PhD61; Ioana Croitoru60; Anne Grete Kristiansen62; Maria Stamelou, MD, PhD63; Helen Mejia Santana64.
Industry and Scientific Advisory Board
Robert Joe Mather35; Minhua Yang, MS36; Christine Brand, MPA37; Cindy Zadikoff38; Kirsten Taylor, PhD40; Thomas McAvoy, PhD41; Baris Bingol, PhD42; Alastair D. Reith, PhD43; Peggy Taylor, ScD44; Gabrielle Ahlberg Hillert, MD PhD45; Mark Minton, MD46; Pierandrea Muglia, PhD47; Robert Umek, PhD48; Barbara Saba50; Lawrence Severt, MD PhD51; Karl Schmidt, PhD52; Matt Troyer, MD53; Kevin Biglan, MD MPH54; Tanya Fischer, MD PhD55; Jeffrey Sevigny, MD56; Adam Schwarz, PhD57; William Marks, MD58; Omar Khwaja, MD PhD59.
Affiliations
1 Institute for Neurodegenerative Disorders, New Haven, CT.
2 The Parkinson’s Institute, Sunnyvale, CA.
3 Northwestern University, Chicago, IL.
4 University of Iowa, Iowa City, IA.
5 Clinical Trials Coordination Center, University of Rochester, Rochester, NY.
6 The Michael J. Fox Foundation for Parkinson’s Research, New York, NY.
7 Innsbruck Medical University, Innsbruck, Austria.
8 Paracelsus-Elena Klinik, Kassel, Germany.
9 University of California, San Francisco, CA.
10 Laboratory of Neuroimaging (LONI), University of Southern California.
11 BioRep, Milan, Italy.
12 University of Pennsylvania, Philadelphia, PA.
13 National Institute on Aging, NIH, Bethesda, MD.
14 Mount Sinai Beth Israel, New York, NY.
15 Indiana University, Indianapolis, IN.
16 Emory University of Medicine, Atlanta, GA.
17 Oregon Health and Science University, Portland, OR.
18 University of Alabama at Birmingham, Birmingham, AL.
19 University of South Florida, Tampa, FL.
20 Baylor College of Medicine, Houston, TX.
21 University of Washington, Seattle, WA.
22 Boston University, Boston, MA.
23 University of Rochester, Rochester, NY
24 Banner Research Institute, Sun City, AZ.
25 Cleveland Clinic, Cleveland, OH.
26 University of Tuebingen, Tuebingen, Germany.
27 University of California, San Diego, CA.
28 Johns Hopkins University, Baltimore, MD.
29 Imperial College of London, London, UK.
30 University of Salerno, Salerno, Italy.
31 Parkinson’s Disease and Movement Disorders Center, Boca Raton, FL.
32 University of Cincinnati, Cincinnati, OH.
33 Macquarie University, Sydney Australia.
34 Hospital Clinic of Barcelona, Barcelona, Spain.
35 Pfizer, Inc., Groton, CT.
36 Biogen, Cambridge, MA.
37 GE Healthcare, Princeton, NJ.
38 AbbVie, Abbot Park, IL.
39 University Hospitals Pitié Salpêtrière, Paris, France.
40 F. Hoffmann La-Roche, Basel, Switzerland.
41 Merck & Co., North Wales, PA.
42 Genentech, Inc., South San Francisco, CA.
43 GlaxoSmithKline, Stevenage, United Kingdom.
44 BioLegend, San Diego, CA.
45 Lundbeck, Copenhagen, Denmark .
46 Avid Radiopharmaceuticals, Philadelphia, PA.
47 UCB Pharma S.A., Brussels, Belgium.
48 Meso Scale Discovery.
49 Piramal Life Sciences, Berlin, Germany.
50 Servier.
51 Allergan, Dublin, Ireland.
52 Celgene, Summit, NJ.
53 Denali Therapeutics, South San Francisco, CA.
54 Eli Lilly and Company, Indianapolis, IN.
55 Sanofi, Paris, France.
56 Prevail Therapeutics, New York, NY.
57 Takeda Pharmaceutical Company, Tokyo, Japan.
58 Verily, South San Francisco, CA.
59 Voyager Therapeutics, Cambridge, MA.
60 Hospital Universitario Donostia, San Sebastian, Spain.
61 Tel Aviv Sourasky Medical Center, Tel Aviv, Israel.
62 St. Olav’s University Hospital, Trondheim, Norway.
63 National and Kapodistrian University of Athens, Athens, Greece.
64 Columbia University Irving Medical Center, New York, NY.
65 Stanford University, Stanford, CA
66 Blackfynn, Philadelphia, PA
67 University of Pittsburgh, Pittsburgh, PA
68 Vincere Biosciences, Inc., Cambridge, MA
69 Center for Strategy Philanthropy at Milken Institute, Washington D.C.
70 University of Medicine and Dentistry of New Jersey, Piscataway, New Jersey




