Membrane-Bound Superoxide Oxidase
Abstract
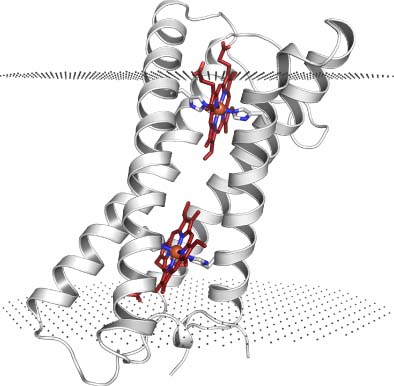
Reactive oxygen species (ROS) are formed as natural byproducts of aerobic metabolism. Their reactivity toward lipids, proteins, and DNA causes extensive cellular damage, and most cells have dedicated enzymes for their scavenging, such as superoxide dismutase (SOD) and catalase. Superoxide oxidase (SOO) is a 20-kDa integral membrane enzyme/protein that directly oxidizes superoxide, reducing ubiquinone in the process. The enzymatic reaction of SOO is diffusion limited, similar to the soluble SOD enzymes. The X-ray structure shows a bundle of four transmembrane (TM) helices with two internal hemes. One heme is partially exposed to the bulk of the periplasm, sitting at the bottom of a positively charged funnel-shaped pocket, presumably representing the entry point of superoxide. The other heme is positioned next to a membrane-exposed cavity, which is likely binding and orienting the ubiquinone head group for the reduction step of the reaction. The structural architecture of SOO resembles the core four-helix, two-heme motif of a group of other diheme enzymes catalyzing a variety of redox reactions. This article includes a detailed description of the structural features of SOO, as well as a discussion on the currently available evidence for its function.