Abstract
Ar.thro.bac'ter. Gr. n. arthron a joint; N.L. masc. n. bacter a rod; N.L. masc. n. Arthrobacter a jointed rod.
Actinobacteria / Actinobacteria / Micrococcales / Micrococcaceae / Arthrobacter
The majority of species exhibit a marked rod–coccus growth cycle when grown in complex media; stationary-phase cultures (generally after 2–7 d) are composed entirely or largely of coccoid cells that are 0.6–1.0 µm in diameter; some species are showing only spherical cells throughout the growth cycle. Fatty acids are predominantly iso- and anteiso-branched with C15:0 anteiso, C15:0 iso, C17:0 anteiso, and C16:0 iso predominating. Few species exhibit significant amounts of C16:0. Quinone system is composed of completely unsaturated or mono-saturated menaquinones with chain lengths of eight to ten isoprenoic units. The cell-wall peptidoglycan contains the diagnostic diamino acid lysine with several variations in the interpeptide bridge conforming to peptidoglycan type A3α or A4α.
DNA G+C content (mol%): 55–72.
Type species: Arthrobacter globiformis (Conn 1928) Conn and Dimmick 1947, 301AL.
The majority of species exhibit a marked rod–coccus growth cycle when grown in complex media; stationary-phase cultures (generally after 2–7 d) are composed entirely or largely of coccoid cells that are 0.6–1.0 µm in diameter; some species are showing only spherical cells throughout the growth cycle. Fatty acids are predominantly iso- and anteiso-branched with C15:0 anteiso, C15:0 iso, C17:0 anteiso, and C16:0 iso predominating. Few species exhibit significant amounts of C16:0. Quinone system is composed of completely unsaturated or mono-saturated menaquinones with chain lengths of eight to ten isoprenoic units. The cell-wall peptidoglycan contains the diagnostic diamino acid lysine with several variations in the interpeptide bridge conforming to peptidoglycan type A3α or A4α.
DNA G+C content (mol%): 55–72.
Type species: Arthrobacter globiformis (Conn 1928) Conn and Dimmick 1947, 301AL.
Number of validated species: 63
Further descriptive information
The genus name Arthrobacter was proposed by Conn and Dimmick (1947), reviving the old name which had been proposed by Fischer (1895). This name was a nomen nudum (naked name) because no species was named, and subsequently the name had been abandoned even by Fischer. Conn and Dimmick (1947) classified three species in the genus Arthrobacter, the type species of the genus Arthrobacter globiforme, Arthrobacter helvolum, and Arthrobacter tumescens. Skerman et al. (1980) then listed Arthrobacter globiforme as Arthrobacter globiformis, and since then this name has been in use. Later, two species of the genus were reclassified in new genera. Arthrobacter tumescens first was classified in the genus Pimelobacter as Pimelobacter tumescens (Suzuki and Komagata, 1983) and later as the type species of the genus Terrabacter, Terrabacter tumescens (Collins et al., 1989). Arthrobacter helvolum was described as the type species of the genus Pseudoclavibacter as Pseudoclavibacter helvolum (Manaia et al., 2004).
Arthrobacters have been recovered from a huge variety of environments, including soils, sea water, fresh water, human skin, oil, brine, tobacco leaves, air, sewage and activated sludge, mural paintings, clinical specimens, and a cyanobacterial mat. van Waasbergen et al. (2000) even detected Arthrobacter strains in terrestrial subsurface sediments at depths between 170 and 220 m.
In the previous edition of Bergey's Manual of Systematic Bacteriology, Keddie et al. (1986) subdivided the genus Arthrobacter into two groups, the Arthrobacter globiformis/Arthrobacter citreus group and the Arthrobacter nicotianae group. Common characteristics of the Arthrobacter globiformis/Arthrobacter citreus group were the presence of the menaquinone MK-9(H2) system and peptidoglycan type A3α, with the characteristic diamino acid L-lysine and an interpeptide chain consisting of monocarboxylic L-amino acids, glycine, or both (Schleifer and Kandler, 1972). Characteristics of the Arthrobacter nicotianae group were menaquinones with completely unsaturated isoprenoic side chains (MK-8 or MK-9) and peptidoglycan type A4α, with L-lysine and an interpeptide bridge containing a dicarboxylic amino acid. Species assigned to the Arthrobacter globiformis/Arthrobacter citreus group were Arthrobacter globiformis, Arthrobacter pascens, Arthrobacter oxydans, Arthrobacter histidinolovorans, Arthrobacter ureafaciens, Arthrobacter ramosus, Arthrobacter ilicis, Arthrobacter aurescens, Arthrobacter citreus, Arthrobacter crystallopoietes, and Arthrobacter atrocyaneus (Kuhn and Starr, 1960; since reclassified as Sinomonas atrocyanea Zhou et al., 2009). Species assigned to the Arthrobacter nicotianae group were Arthrobacter nicotianae, Arthrobacter protophormiae, Arthrobacter uratoxydans, and Arthrobacter sulfureus. Due to the absence of chemotaxonomic data, several other species were treated as species incertae sedis, including Arthrobacter mysorens (phylogenetically this species is related to the Arthrobacter nicotianae group; Figure 1); Arthrobacter picolinophilus [according to Koch et al. (1995) this species is a later heterotypic synonym of Rhodococcus erythropolis], Arthrobacter radiotolerans [reclassified as Rubrobacter radiotolerans (Suzuki et al., 1988)]; and Arthrobacter siderocapsulatus [according to Chun et al. (2001), this species is a later heterotypic synonym of Pseudomonas putida]. Other species showing meso- or LL-diaminopimelic acid as the characteristic diamino acid in the peptidoglycan were listed in two addenda: Arthrobacter duodecanis [reclassified as Tetrasphaera duodecanis (Ishikawa and Yokota, 2006)]; Arthrobacter variabilis [reclassified as Corynebacterium variabilis (Collins, 1987) and then renamed Corynebacterium variabile (Euzéby, 1998)]; Arthrobacter viscosus (the type of this species shares 96.9–99.6% similarity with species of the genus Rhizobium and hence reclassification as a species of Rhizobium is desirable); Arthrobacter simplex [reclassified as Pimelobacter simplex (Suzuki and Komagata, 1983)]; and Arthrobacter tumescens [reclassified as Terrabacter tumescens (Collins et al., 1989)].
Based on the amino acid composition of the peptidoglycan interpeptide chain, another grouping of Arthrobacter species was proposed by Komagata and Suzuki (1987) which dissected Arthrobacter species into seven groups designated Komagata/Suzuki groups I–VII (Table 1).
| Komagata/Suzuki group | Amino acid composition of the interpeptide bridge | Species |
|---|---|---|
| I | Lys–Ser–Thr–Ala | A. oxydans, A. polychromogenes |
| II | Lys–Ala–Thr–Ala | A. aurescens, A. histidinolovorans, A. ilicis, A. nicotinovorans, A. ureafaciens |
| III | Lys–Ala1–4 | A. crystallopoietes, A. globiformis, A. pascens, A. ramosus |
| IV | Lys–Ser–Ala2–3 | Sinomonas atrocyanea (formerly A. atrocyaneus) |
| V | Lys–Thr–Ala2 | A. citreus |
| VI | Lys–Ala–Glu | A. nicotianae, A. creatinolyticus, A. uratoxydans, A. protophormiae |
| VII | Lys–Glu | A. sulfureus |
- a Abbreviations: Lys, lysine; Ser, serine; Thr, threonine; Ala, alanine; Glu, glutamic acid.
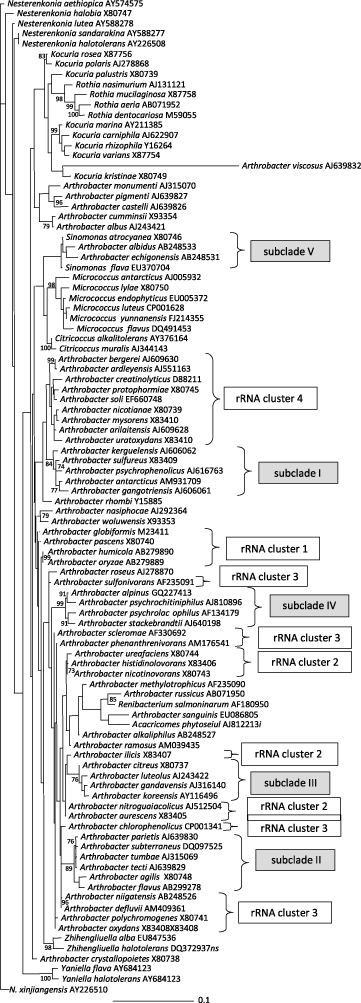
At present, the genus Arthrobacter consists of 64 recognized species. Based upon the EzTaxon server (Chun et al., 2007), with the exception of Arthrobacter viscosus, these Arthrobacter species share 16S rRNA gene sequence similarities of 94.4–99.3% with the type strain of the genus, Arthrobacter globiformis. Several species of related genera such as Acaricomes, Brachybacterium, Citricoccus, Kocuria, Micrococcus, Nesterenkonia, Rothia, Sinomonas, and Zhihengliuella also possess sequence similarities with Arthrobacter globiformis within this range. These observations indicate the heterogeneity of the genus Arthrobacter. Hence, identification of novel strains at the genus level only based on 16S rRNA gene sequence similarities are ambiguous if a high degree of similarity is not found with Arthrobacter globiformis. Also phylogenetic examinations indicate that Arthrobacter is a polyphyletic genus. Several Micrococcaceae genera, such as Micrococcus, Citricoccus, Sinomonas, Acaricomes, and Renibacterium, are included in the clade comprising all Arthrobacter species and separate the genus into several subclades (Figure 1). However, the majority of branching points in the phylogenetic tree calculated applying the maximum-likelihood (Figure 1) or neighbor-joining algorithms (not shown) are not supported by significant boot-strap values (>75%), indicating that the phylogenetic relationships among numerous Arthrobacter species remain ambiguous at this time.
Species of the Arthrobacter globiformis/Arthrobacter citreus group (Keddie et al., 1986) are distributed throughout the phylogenetic tree. Although not supported by significant bootstrap values, in many trees Arthrobacter globiformis and Arthrobacter pascens appear on a common branch, and Arthrobacter ureafaciens and Arthrobacter histidinolovorans appear on another (Figure 1). The remaining species of the Arthrobacter globiformis/Arthrobacter citreus group are distributed throughout phylogenetic trees and do not appear to be closely related to each other. For this reason, this group is probably artificial, and it is not used further in this chapter. In contrast, species assigned to the Arthrobacter nicotianae group form a distinct clade separate from other Arthrobacter species but without significant bootstrap support.
In the tree consisting of all Arthrobacter species and representatives of all genera assigned to the family Micrococcaceae, five subclades (which may be considered as genus-like clusters) can be identified. These subclades all have at least moderate bootstrap support. Subclade I is composed of Arthrobacter antarcticus, Arthrobacter gangotriensis, Arthrobacter kerguelensis, Arthrobacter psychrophenolicus, and Arthrobacter sulfureus with 84% bootstrap support. Since this subclade contains Arthrobacter sulfureus, the only representative of Komagata/Suzuki group VII, the other four species may also be assigned to this group. In fact, the peptidoglycan types of Arthrobacter antarcticus, Arthrobacter gangotriensis, Arthrobacter kerguelensis, and Arthrobacter psychrophenolicus support this assignment. Subclade II comprises six species, Arthrobacter agilis, Arthrobacter flavus, Arthrobacter parietis, Arthrobacter subterraneus, Arthrobacter tecti, and Arthrobacter tumbae with 89% bootstrap support. Subclade III is formed by Arthrobacter citreus (originally assigned to the Arthrobacter globiformis/Arthrobacter citreus group Keddie et al., 1986) or Komagata/Suzuki group V (Komagata and Suzuki, 1987), Arthrobacter luteolus, Arthrobacter gandaviensis, and Arthrobacter koreensis with 76% bootstrap support. A very stable subclade IV with 99% bootstrap support consists of Arthrobacter alpinus, Arthrobacter psychrochitiniphilus, Arthrobacter psychrolactophilus, and Arthrobacter stackebrandtii. Subclade V (100% bootstrap support) is composed of the two described Sinomonas species Sinomonas flava and Sinomonas atrocyanea (formerly named Arthrobacter atrocyaneus and assigned to Arthrobacter globiformis/Arthrobacter citreus group and Komagata/Suzuki group IV (Zhou et al., 2009), Arthrobacter albidus, and Arthrobacter echigonensis. The type species of the genus, Arthrobacter globiformis, is not assigned to any of these sub-clades or any other groups of species whose relatedness is supported by high bootstrap values (>75%), including Arthrobacter humicola/Arthrobacter oryzae, Arthrobacter pigmenti/Arthrobacter castelli, Arthrobacter defluvii/Arthrobacter niigatensis, Arthrobacter bergerei/Arthrobacter ardleyensis, and Arthrobacter albus/Arthrobacter cumminsii.
Several groups composed of species sharing high similarities (>97%) in their 16S rRNA gene sequences can also be identified. These rRNA similarity groups are not confirmed upon phylogenetic analyses and are here designated rRNA clusters, which distinguished them from the subclades. rRNA cluster 1 comprises Arthrobacter globiformis, Arthrobacter pascens, Arthrobacter humicola, and Arthrobacter oryzae, which share 98.4–99.5% sequence similarity. Two species of this cluster are also classified in the Arthrobacter globiformis/Arthrobacter citreus group and Komagata/Suzuki group III. Species assigned to rRNA cluster 2 are Arthrobacter histidinolovorans, Arthrobacter nicotinovorans, Arthrobacter ureafaciens, Arthrobacter ilicis, Arthrobacter aurescens, and Arthrobacter nitroguajacolicus; and sequence similarities are in the range 97.6–99.7%. This cluster contains species of the Arthrobacter globiformis/Arthrobacter citreus group and all species assigned to Komagata/Suzuki group II. rRNA cluster 3 comprises Arthrobacter chlorophenolicus, Arthrobacter sulfonivorans, Arthrobacter oxydans, Arthrobacter defluvii, Arthrobacter polychromogenes, Arthrobacter scleromae, Arthrobacter niigatensis, and Arthrobacter phenanthrenivorans, with sequence similarities in the range of 97.3–99.7%. The cluster contains one species assigned to the Arthrobacter globiformis/Arthrobacter citreus group and two species assigned to Komagata/Suzuki group I. rRNA cluster 4 species share 96.7–99.7% sequence similarity. This cluster contains the species Arthrobacter arilaitensis, Arthrobacter ardleyensis, Arthrobacter bergerei, Arthrobacter creatinolyticus Arthrobacter mysorens, Arthrobacter nicotianeae, Arthrobacter protophormiae, Arthrobacter soli, and Arthrobacter uratoxydans. Three of these species were originally in the Arthrobacter nicotianae group and Komagata/Suzuki group VI. Notably, Arthrobacter bergerei shares high sequence similarity of 96.6–97.0% with some subclade I species, and subclade I species Arthrobacter antarcticus, Arthrobacter psychrophenolicus, and Arthrobacter kerguelensis also share sequence similarity values in the range 96.7–97.1% with some rRNA cluster 4 species.
In a study on genetic diversity among Arthrobacter species collected from terrestrial deep-subsurface sediments, partial recA gene sequences were obtained from strains of 14 Arthrobacter species (van Waasbergen et al., 2000). Unfortunately, in this study only 360 nucleotides of each strain were analyzed, and hence the deduced phylogeny is of limited statistical significance. However, these phylogenetic analyses demonstrated a close relationship between these groups of Arthrobacter species, Arthrobacter sulfureus/Arthrobacter uratoxydans/Arthrobacter protophormiae/Arthrobacter nicotianae, Arthrobacter pascens/Arthrobacter globiformis, Arthrobacter aurescens/Arthrobacter nicotinovorans/Arthrobacter histidinolovorans/Arthrobacter ureafaciens, and Arthrobacter oxydans/Arthrobacter polychromogenes, supporting the 16S rRNA based phylogeny. The low sequence similarity of the Arthrobacter agilis and Arthrobacter citreus genes to those from the other species also agrees with the findings of 16S rRNA studies. Interestingly, two species of Micrococcus, Micrococcus luteus and Micrococcus lylae, were clearly distinguished from all of the Arthrobacter species examined. In contrast, the 16S rRNA gene trees do not clearly distinguish these species from Arthrobacter (Figure 1). In other studies, the recA gene phylogeny also suggests a high degree of relatedness between Arthrobacter phenanthrenivorans, Arthrobacter oxydans, and Arthrobacter polychromogenes, which is similar to the 16S rRNA phylogeny (Kallimanis et al., 2009). Lastly, on the basis of its recA sequence, Arthrobacter chlorophenolicus does not appear to be closely related to other Arthrobacter species.
Chemotaxonomy
Fatty acid profiles of Arthrobacter species are composed predominantly of iso- and anteiso-branched fatty acids. The majority of species exhibit fatty acid profiles dominated by C15:0 anteiso and possessing usually high amounts of C15:0 iso, C16:0 iso, and C17:0 anteiso. Several species also possess significant amounts of C16:0, including Arthrobacter citreus, Arthrobacter nicotianae, Arthrobacter cumminsii, Arthrobacter niigatensis, Arthrobacter oxydans, and Arthrobacter polychromogenes. However, the presence of this fatty acid is not correlated with genetic relatedness. Moreover, Arthrobacter russicus possesses an unusual fatty acid profile dominated by C17:0 anteiso, but this trait has not been reported in its closest relative Renibacterium salmonniarum, which also possesses large amounts of C15:0 anteiso (Embley et al., 1983). Therefore, while the fatty acid data may be useful for characterization and identification of Arthrobacter species, it does not correlate well with phylogenetic relationships between closely related species.
Arthrobacter species can be distinguished into two major groups based on the presence of monosaturated or completely unsaturated quinones. The first group includes the majority of Arthrobacter species, which possess monosaturated menaquinone with nine isoprenoic units in the side chain, MK-9(H2). This group also includes a smaller number of species with predominantly MK-8(H2) and corresponds to the polyphyletic Arthrobacter globiformis/Arthrobacter citreus group of Keddie et al. (1986). The second group includes several Arthrobacter species that exhibit menaquinones with eight, nine, and/or ten completely unsaturated isoprenoic units (MK-8, MK-9, and MK-10) and corresponds to the Arthrobacter nicotianae group (Keddie et al., 1986). Since the designation of these groups, all newly proposed Arthrobacter species with completely unsaturated menaquinones have proven to be phylogenetically related to the Arthrobacter nicotianae group. Species of the Arthrobacter nicotianae group (Keddie et al., 1986) whose menaquinone system have been analyzed include Arthrobacter nicotianae, Arthrobacter creatinolyticus, Arthrobacter protophormiae, Arthrobacter sulfureus, Arthrobacter gangotriensis, Arthrobacter psychrophenolicus, Arthrobacter antarcticus, and Arthrobacter kerguelensis. Unfortunately, the menaquinones of many related species of this group, such as Arthrobacter arilaitensis, Arthrobacter bergerei, Arthrobacter mysorens, Arthrobacter rhombi, and Arthrobacter soli, have not been examined. However, it would not be surprising if these latter species also possess completely unsaturated quinones.
Several Arthrobacter species exhibit quinone systems unusual for the genus. Arthrobacter koreensis (subclade III) was reported to have a quinone system with almost equal amounts (43:35) of MK-8(H2) and MK-9(H2). Other members of subclade III, Arthrobacter luteolus and Arthrobacter citreus, were reported to contain a different ratio of MK-8(H2) and MK-9(H2), 22:68 and 30:67, respectively (Collins and Kroppenstedt, 1983; Lee et al., 2003), which might reflect their close relatedness. Another quinone system with also almost equal amounts of MK-9(H2) and MK-8(H2), a ratio of 56:44, was reported for Arthrobacter nasiphocae (Collins et al., 2002a), which was not placed in any of the subclades above. Arthrobacter albus and Arthrobacter cumminsii, which are phylogenetically quite distant from the type species of the genus (Figure 1), exhibit the major menaquinone MK-8(H2) (Busse, unpublished results). Species of subclade III, Arthrobacter parietis, Arthrobacter subterraneus, Arthrobacter tecti, and Arthrobacter tumbae share a quinone system with largely MK-9(H2) and significant amounts (>20%) of MK-10(H2), but this quinone system is also found in the distant relative Arthrobacter stackebrandtii. A quinone system most unusual among arthrobacters and many other bacteria was reported for Arthrobacter scleromae. It consists of unsaturated and saturated menaquinones of MK-8(H2) and minor amounts of MK-10 (ratio of 88:12) (Huang et al., 2005b). Likewise, the quinone system of Arthrobacter phenanthrenivorans consists of MK-8 and MK-9(H2) in a ratio 3.6:1 (Kallimanis et al., 2009). Because quinone systems composed of monosaturated and completely unsaturated menaquinones with isoprenoic side chains of different lengths are most unusual among bacteria, re-analyses would be desirable to confirm these two quinone systems.
The peptidoglycans of Arthrobacter species contain the diagnostic diamino acid lysine. Applying the scheme of Schleifer and Kandler (1972), differences in the amino acid composition of the interpeptide chains distinguish Arthrobacter species into two major groups. The majority of species exhibit peptidoglycan type A3α, which possess monocarboxylic L-amino acids or glycine or both in the interpeptide bridge. Arthrobacter species with type A3α peptidoglycan can be distinguished based on the amino acid composition in the interpeptide bridge. Arthrobacter species of subclade II and III are characterized by an interpeptide bridge consisting of Lys–Thr–Ala2–3. Subclade II species Arthrobacter agilis, Arthrobacter flavus, Arthrobacter subterraneus, Arthrobacter tecti, and Arthrobacter tumbae possess Lys–Thr–Ala3, while Arthrobacter parietis possesses Lys–Thr–Ala2. Subclade III species (Arthrobacter citreus, Arthrobacter gandavensis, Arthrobacter koreensis, and Arthrobacter luteolus) also possess Lys–Thr–Ala2. Other species, Arthrobacter psychrolactophilus and Arthrobacter alpinus grouped in subclade IV, are also characterized by an interpeptide bridge of Lys–Thr–Ala3. This interpeptide bridge may also be present in Arthrobacter psychrochitiniphilus, whose composition is known to be Lys, Thr, and Ala.
Species of Arthrobacter rRNA cluster 2 (Arthrobacter aurescens, Arthrobacter histidinolovorans, Arthrobacter ilicis, Arthrobacter nicotinovorans, Arthrobacter nitroguajacolicus, and Arthrobacter ureafaciens) exhibit an interpeptide bridge of Lys–Ala–Thr–Ala. Another similar interpeptide bridge (Lys–Ser–Thr–Ala) is found in Arthrobacter chlorophenolicus, Arthrobacter sulfonivorans, Arthrobacter oxydans, Arthrobacter defluvii, Arthrobacter polychromogenes, Arthrobacter scleromae, and probably also Arthrobacter niigatensis and Arthrobacter alkaliphilus. For the latter two species only the predominant amino acids of Lys, Ser, Thr, and Ala were reported (Ding et al., 2009). Except Arthrobacter alkaliphilus, which shares highest 16S rRNA gene similarity with Arthrobacter methylotrophus (97.4%), these species are grouped in rRNA cluster 3. The peptidoglycan of Arthrobacter albidus and Arthrobacter echigonensis is composed of the amino acids Lys, Ser, and Ala (Ding et al., 2009), which is consistent with a peptidoglycan interpeptide bridge of Lys–Ser–Alax and placement of these two species in subclade V together with the species of the genus Sinomonas.
Among arthrobacters, Arthrobacter nasiphocae and Arthrobacter roseus possess a unique interpeptide bridge composed of Lys–Ala2–Gly2–3–Ala (Gly) and Lys–Gly–Ala3, respectively, which might reflect their distant phylogenetic relationship to other Arthrobacter species. The presence of serine in the interpeptide bridge of Arthrobacter castelli (Lys–Ala–Ser–Ala3) distinguishes this species from its closest relative Arthrobacter pigmenti, which possesses Lys–Ala4 in the interpeptide chain. Nevertheless, the presence of four alanine residues in the interpeptide bridges of both Arthrobacter castelli and Arthrobacter pigmenti may support the close relatedness of the two species. Other Arthrobacter species with interpeptide bridges composed of only alanine in addition to lysine are distributed throughout the genus. These species are Arthrobacter crystallopoietes (Lys–Ala), Arthrobacter pascens (Lys–Ala2), Arthrobacter stackebrandtii (Lys–Ala2), Arthrobacter humicola (Lys–Ala>2), Arthrobacter oryzae (Lys–Ala>2), Arthrobacter globiformis (Lys–Ala3), Arthrobacter russicus (Lys–Ala2), Arthrobacter methylotrophus (Lys–Ala2–4), Arthrobacter monumenti (Lys–Ala4), Arthrobacter pigmenti (Lys–Ala4), and Arthrobacter ramosus (Lys–Ala4). Arthrobacter globiformis, Arthrobacter humicola, Arthrobacter oryzae, and Arthrobacter pascens are grouped in rRNA cluster 1, and this grouping is supported by the intepeptide composition. In contrast, 16S rRNA analyses do not suggest assignment of the remaining species to rRNA cluster 1. Instead, relatedness to species with different interpeptide bridge compositions is indicated. Hence, this type of interpeptide bridge appears to be less significant for grouping of Arthrobacter than other interpeptide bridge structures.
The other major peptidoglycan type found in Arthrobacter species is A4α, which contains glutamic acid or glutamic acid and alanine in the interpeptide bridge. It is found in all species in the Arthrobacter nicotianae group (species of subclade I and rRNA cluster 4) and in Arthrobacter rhombi. Many phylogenetic trees place Arthrobacter rhombi at the root of the Arthrobacter nicotianae group, but bootstrap support is lacking for assignment to this group (Figure 1). Nevertheless, the type of interpeptide bridge Lys–Ala–Glu supports its affiliation with rRNA cluster 4. Species of subclade I, Arthrobacter sulfureus, Arthrobacter antarcticus, Arthrobacter gangotriensis, Arthrobacter psychrophenolicus, and Arthrobacter kerguelensis, possess an interpeptide bridge of Lys–Glu. Peptidoglycan type A4α with L-glutamic acid in the interpeptide bridge is also present in Arthrobacter cumminsii and Arthrobacter albus, which together occupy a phylogenetically distinct branch within the radiation of Arthrobacter species (Figure 1). Another species with peptidoglycan type A4α is Arthrobacter woluwensis. In this case, the dicarboxylic acid in the interpeptide bridge is D-aspartic acid, which (so far) is a unique trait within the genus Arthrobacter but this trait was already reported in Micrococcus flavus and a strain of Micrococcus luteus (Liu et al., 2007; Wieser et al., 2002).
Cell-wall sugars have been analyzed for approximately half of Arthrobacter species, and so far 14 different sugar compositions have been reported. The majority of species possess galactose (Gal) solely or in combination with other sugars, such as glucose (Glc), rhamnose (Rha), mannose (Man), ribose (Rib), and xylose (Xyl). Gal alone is present in the cell walls of Arthrobacter tumbae, Arthrobacter parietis, and Arthrobacter citreus. The cell walls of the type species of the genus, Arthrobacter globiformis, and its close relative Arthrobacter pascens contain Gal and Glc, and these sugars are also found in Arthrobacter oryzae, Arthrobacter crystallopoietes, Arthrobacter histidinolovorans, Arthrobacter oxydans, Arthrobacter scleromae, Arthrobacter nicotianae, and Arthrobacter sulfureus. Acaricomes phytoseiuli, which is phylogenetically placed within the radiation of the genus Arthrobacter, also possesses these sugars. Arthrobacter tecti, Arthrobacter aurescens, Arthrobacter alpinus, and Arthrobacter ureafaciens possess Gal and minor amounts of Man. Arthrobacter humicola and Arthrobacter pigmenti possess Gal and Rha in their cell walls. The cell wall of Arthrobacter defluvi contains Gal, Glc, and Rha. The cell walls of Arthrobacter subterraneus and Arthrobacter flavus contain Gal, Glc, and Rib. The cell walls of Arthrobacter ramosus and Arthrobacter ilicis possess Gal, Man, and Rha.The cell walls of Arthrobacter monumenti and Arthrobacter castelli possess minor amounts of Xyl and Rha in addition to Gal. Four cell-wall sugars, Gal, Glc, Rib, and Rha were detected in Arthrobacter roseus, and Gal, Glc, Man, and Rib were found in Arthrobacter arilaitensis. Several species lack Gal in the cell wall. Only one cell-wall sugar was detected in Arthrobacter psychrophenolicus (Glc) and Arthrobacter koreensis and Arthrobacter luteolus (Rha). Arthrobacter agilis is the sole Arthrobacter species which contains glucosamine (GlcN). Three cell-wall sugars, Glc, Man, and Rib, are present in Arthrobacter bergerei. Since the cell-wall sugar composition varies among closely related Arthrobacter species or certain sugar compositions are found in different subclades/rRNA clusters, apparently this feature is more suitable for characterization of species rather than for identification of closely related groups.
In a study of distribution and composition of teichoic acids, Fiedler and Schäfler (1987) showed that these components are found in the cell walls of A4α peptidoglycan type arthrobacters (Arthrobacter nicotianae group sensu Keddie et al., 1986), but not in other representatives of the genus. The teichoic acids of Arthrobacter nicotianae are composed of glycerol, glucose, and glucosamine. A qualitatively similar teichoic acid composition is also present in a strain of Arthrobacter mysorens, whereas Arthrobacter protophormiae and Arthrobacter sulfureus also possess galactosamine. It might be assumed that the teichoic acids contribute to the cell-wall sugars, but surprisingly the glucosamine of the teichoic acids of Arthrobacter nicotianae and Arthrobacter sulfureus was not reported among the cell-wall sugars (Stackebrandt et al., 1983b). Unfortunately, the teichoic acids of only a very limited number of species have been analyzed. Hence, the importance of this tool for classification is unclear.
Information regarding polar lipid composition of arthrobacters is also rather limited. All Arthrobacter species which have been examined possess phosphatidylglycerol and diphosphatidylglycerol. In addition, phosphatidylinositol and several glycolipids may be present. In an early study, Walker and Bastl (1967) identified the glycolipids of Arthrobacter globiformis strain 616 as dimannosyl-diglyceride, monogalactosyl-diglyceride, and digalactosyl-diglyceride. They also found some evidence for the presence of small amounts of a trimannosyl diglyceride and an even smaller quantity of a tetramannosyl-diglyceride. In a following study, Shaw and Stead (1971) analyzed the lipid composition of the type strains of Arthrobacter crystallopoietes and Arthrobacter pascens as well as Arthrobacter globiformis strain 616. The polar lipid profile of Arthrobacter crystallopoietes was composed of phosphatidylglycerol, diphosphatidylglycerol, phosphatidylinositol, dimannosyl-diglyceride, monogalactosyl-diglyceride, digalactosyl-diglyceride, and a fourth glycolipid supposed to be tetramannosyl-diglyceride. No significant differences were reported for Arthrobacter pascens and Arthrobacter globiformis. These data confirm the close phylogenetic relatedness of Arthrobacter globiformis and Arthrobacter pascens and may imply relatedness to Arthrobacter crystallopoietes, which does not occupy a stable position in phylogenetic trees. The polar lipid profile of Arthrobacter ilicis consists of phosphatidylglycerol, diphosphatidylglycerol, phosphatidylinositol, and two unidentified glycolipids GA and GB (Collins et al., 1981). The polar lipids of the type strains of Arthrobacter aurescens, Arthrobacter citreus, Arthrobacter crystallopoietes, Arthrobacter globiformis, and Arthrobacter polychromogenes also comprise phosphatidylglycerol, diphosphatidylglycerol, phosphatidylinositol, and varying numbers of unidentified glycolipids (Collins et al., 1982a). The number of Arthrobacter globiformis glycolipids (although not analyzed for chemical structure) was in agreement with the number in previous reports. Collins et al. (1982a) detected only three glycolipids in Arthrobacter cyrstallopoietes, but already Shaw and Stead (1971) mentioned the low amounts of a fourth glycolipid in this species. Arthrobacter aurescens and Arthrobacter polychromogenes showed three unidentified glycolipids, whereas in Arthrobacter citreus only two glycolipids were detected.
Collins and Kroppenstedt (1983) investigated the polar lipid profiles of the type strains of Arthrobacter citreus, Arthrobacter nicotianae, Arthrobacter protophormiae, Arthrobacter sulfureus (formerly Brevibacterium sulfureus) and found polar lipid profiles consisting of phosphatidylglycerol, diphosphatidylglycerol, and unidentified glycolipids, either Ga or Gb. These two glycolipids showed chromatographic behaviors of diglycosyldiacylglycerol, and their staining behavior indicated the presence of galactose and/or mannose residues. Unidentified glycolipid Ga was detected in Arthrobacter nicotianae, Arthrobacter protophormiae, and Arthrobacter citreus, whereas Gb was found in Arthrobacter sulfureus. Arthrobacter citreus was the only species of this group which possessed phosphatidylinositol. The distribution of Ga and Gb among species of the Arthrobacter nicotianae group, here represented by Arthrobacter nicotianae, Arthrobacter protophormiae and Arthrobacter sulfureus, is in accordance with the subdivision of this group based on phylogeny and peptidoglycan type (Lys–Ala–Glu or Lys–Glu). The lipids of Arthrobacter psychrophenolicus, placed with Arthrobacter sulfureus in subclade I (Figure 1), are phosphatidylglycerol, diphosphatidylglycerol, phosphatidylinositol, and an unidentified glycolipid (Margesin et al., 2004). Unfortunately, no information is provided concerning the chromatographic behavior of this unidentified glycolipid which would allow a comparison to Gb. However, the presence of phosphatidylinositol distinguishes Arthrobacter psychrophenolicus from its close relative Arthrobacter sulfureus. The unidentified glycolipid GA of Arthrobacter ilicis shows a chromatographic behavior similar to Ga mentioned above, whereas the chromatographic behavior of GB of Arthrobacter ilicis suggests that it is different from Gb. Arthrobacter russicus is characterized by the predominant lipids diphosphatidylglycerol and phosphatidylinositol and lesser amounts of phosphatidylglycerol (Li et al., 2004c). In addition to possessing phosphatidylglycerol, diphosphatidylglycerol, and phosphatidylinositol, Arthrobacter castelli, Arthrobacter monumenti and Arthrobacter pigmenti possess one unidentified phospholipid, and one unidentified glycolipid; Arthrobacter parietis possesses one unidentified phospholipid and two unidentified glycolipids; and Arthrobacter tecti and Arthrobacter tumbae possess one unidentified phospholipid (Heyrman et al., 2005). Arthrobacter flavus, Arthrobacter roseus, and Arthrobacter phenanthrenivoras were reported to possess phosphatidylglycerol, diphosphatidylglycerol, and phosphatidylethanolamine (Kallimanis et al., 2009; Reddy et al., 2000; Reddy et al., 2002). Phosphatidylethanolamine is rare among species of Arthrobacter and related genera as is the absence of phosphatidylinositol. Re-analysis of the polar lipid profile did not provide any indication of the presence of phosphatidylethanolamine in the type strain of Arthrobacter roseus. Like other arthrobacters, the type strain of Arthrobacter roseus had a polar lipid profile containing predominantly diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol, and an unidentified glycolipid (Busse, unpublished results). Nevertheless, the variability in polar lipid composition suggest that, while this trait is useful for classification of arthrobacters, current data are not complete enough to demonstrate that the distribution of polar lipids is dependent on phylogenetic relatedness.
Information concerning polyamine patterns, which are often useful for classification of bacteria, is limited. Altenburger et al. (2002) analyzed polyamines of 13 Arthrobacter species, but the results were not very useful for classification. Spermidine was the most abundant polyamine in most species. While significant differences in the overall polyamine content were detected, the polyamine content was not in accordance with relationships deduced from phylogeny or other chemotaxonomic markers.
Pathogenicity
Although soil is considered the major habitat of arthrobacters, strains have been also isolated from clinical specimens and may have been previously identified as CDC coryneform group B-1 and B-3 (Funke et al., 1997b). Thus, several Arthrobacter strains now have been reported to be associated with human diseases.
Arthrobacter cumminsii strains were isolated from urine, a skin infection (Funke et al., 1996), a urinary tract infection, chronic otorrhea, infected amniotic fluid, a vaginal swab, calcaneus osteomyelitis, external otitidis, a deep tissue infection of the upper leg, chronic cervicitis, a blood culture, and a leg wound (Funke et al., 1998). A strain of Arthrobacter woluwensis was isolated from blood of an HIV-infected patient hospitalized with fever and chills (Funke et al., 1996) and found to cause a subacute infective endocarditis with involvement of the native mitral valve of an HIV-seronegative injection drug user (Bernasconi et al., 2004) and a catheter-related bacteremia (Shin et al., 2006). Wauters et al. (2000a) reported isolation and characterization of the newly described species Arthrobacter luteolus (one strain) and Arthrobacter albus (two strains) from an infected surgical wound, and urine and blood from a patient with severe phlebitis, respectively, and detected Arthrobacter oxydans (two strains) in blood specimens. The species Arthrobacter creatinolyticus was identified in a screening for creatinine-hydrolyzing bacteria from patients with unusually low levels of creatinine in urine samples (Hou et al., 1998), and Arthrobacter scleromae was isolated from bloody effusion of a swollen scleroma (Huang et al., 2005b). A bacterial strain associated with a case of Whipple's syndrome in a patient with severe chronic uveitis and systemic inflammatory manifestations was classified within the genus Arthrobacter based on partial 16S rRNA gene sequence comparison (Bodaghi et al., 1998). Unnamed Arthrobacter strains were recovered from blood cultures or reported to be associated with vaginitis and endophthalmitis (Esteban et al., 1996; Funke et al., 1996). Another unnamed Arthrobacter strain was isolated from the blood of a neutropenic patient with acute lymphoblastic leukemia (Hsu et al., 1998).
Among 50 strains of large-colony-forming, whitish-grayish, non-cheese-like-smelling, nonfermentative Gram-stain-positive rods encountered from human clinical specimens, Mages et al. (2008) identified 38 strains of the genus Arthrobacter. Fourteen strains isolated from blood, urine, otitis externa, cervix, wound swab, and a tracheal secretion were close relatives of Arthrobacter cumminsii. Eleven strains isolated from a blood culture, lung swab after autopsy, eye, vaginal swab, wound swab, or unknown source were closely related to Arthrobacter oxydans.
Close relatives of Arthrobacter aurescens were recovered from urine and an unknown clinical source. One strain isolated from neck abscess was affiliated with Arthrobacter oryzae, two strains related to Arthrobacter albus were isolated from blood culture and urine, one strain related to Arthrobacter protophormiae was isolated from urine, and some strains with unclear species affiliations were present in specimens from various blood cultures and a hand wound. The novel species Arthrobacter sanguinis was also isolated from blood (Mages et al., 2008).
These studies demonstrate that arthrobacters may cause disease in humans and that at least the species Arthrobacter cumminsii and Arthrobacter oxydans are opportunistic pathogens. Moreover, the pathogenic potential of Arthrobacter albus, Arthrobacter woluwensis, Arthrobacter arborescens, Arthrobacter oryzae, Arthrobacter protophormiae, and Arthrobacter sanguinis should be studied in more detail.
Arthrobacters are generally sensitive to antibiotics. However, only few reports exist on the susceptibility of arthrobacters to antibiotics. Among 24 Arthrobacter strains (including the type strains of Arthrobacter globiformis, Sinomonas atrocyanea [formerly Arthrobacter atrocyaneus], Arthrobacter aurescens, Arthrobacter crystallopoietes, Arthrobacter cumminsii, Arthrobacter histidinolovorans, Arthrobacter nicotinovorans, Arthrobacter oxydans, Arthrobacter pascens, Arthrobacter ureafaciens, Arthrobacter nicotianeae, Arthrobacter protophormiae, Arthrobacter uratoxydans, and Arthrobacter woluwensis) tested for susceptibility to 16 antimicrobials (amoxicillin-clavulanic acid, ampicillin, ceftriaxone, cefuroxime, cefalothin, chloramphenicol, ciprofloxacin, clindamycin, erythromycin, gentamicin, imipenem, penicillin G, rifampin, teichoplanin, tetracycline, and vancomycin), only the strain of Arthrobacter woluwensis was multidrug resistant. It was only susceptible to teichoplanin, tetracycline, and vancomycin (Funke et al., 1996). The susceptibility patterns of 38 Arthrobacter strains was also studied by Mages et al. (2008). Nearly all strains were susceptible to β-lactams, doxycycline, gentamicin, linezolid, rifampin, and vancomycin. Additional strains of Arthrobacter woluwensis examined in the latter study were not multidrug resistant, indicating that this trait was not characteristic of the species.
Differentiation of the genus Arthrobacter from other genera
Differentiation of the genus Arthrobacter from distantly related genera which share phenotypic properties that could lead to misidentifications (e.g. genera of the family Microbacteriaceae, Brevibacterium, or Corynebacterium) can be unambiguously achieved by 16S rRNA gene sequence analyses. Support for phylogenetic relationshhips based on 16S rRNA analyses may be obtained from analyses of the peptidoglycan type, quinone system, and polyamines. Members of the family Microbacteriaceae possess exclusively B type peptidoglycan, usually containing ornithine, diaminobutyric acid, or lysine as the diagnostic diamino acid. Completely unsaturated menaquinones with 9–14 isoprenoic units are also useful for differentiation from the majority of Arthrobacter species. The polyamine pattern of Arthrobacter differentiates it from certain genera, such as Agrococcus, Brevibacterium, and Pseudoclavibacter helvolus, whose polyamine patterns are predominated by spermine, putrescine, or cadaverine (Altenburger et al., 1997; Manaia et al., 2004; Wieser et al., 1999; Zlamala et al., 2002). Differentiation of Arthrobacter from other genera of the Micrococcaceae is hampered because Arthrobacter is not monophyletic and possesses significant phenotypic heterogeneity, especially for chemotaxonomic markers such as the interpeptide chains in the peptidoglycan and quinone systems. Hence, 16S rRNA based phylogeny and sequence similarities do not unambiguously allow identification at the genus level.
The majority of Arthrobacter species, except Arthrobacter cumminsii, Arthrobacter albus, and Arthrobacter scleromae, can be distinguished from members of the genus Micrococcus by the predominance of MK-7(H2), MK-8, and/or MK-8(H2) in the latter genus. Except for Arthrobacter woluwensis, which contains L-Lys–D-Asp, no Arthrobacter species has a peptidoglycan interpeptide bridge similar to that of Micrococcus species, which consists of L-Lys–D-Asp or L-Lys–polymerized peptide subunit (subgroup A2; Schleifer and Kandler, 1972). The only representative of the genus Acaricomes contains a MK-10(H2) quinone system, which has been reported in moderate or low amounts in only a few Arthrobacter species.
The MK-9(H2) quinone system of Citricoccus is common in the majority of Arthrobacter species, but it distinguishes Citricoccus from Arthrobacter species with completely unsaturated menaquinones. The peptidoglycan of Citrobacter species contains an interpeptide chain composed of Lys–Gly–Glu, which is not known for Arthrobacter, and allows a reliable differentiation between the two genera. Kocuria species form a rather stable rRNA gene clade within the Micrococcaceae, and phylogenetic analyses can differentiate this genus. The majority of Arthrobacter species are also distinguished from Kocuria based on the amino acid composition of the interpeptide bridge, which consists of Lys–Ala3–4 in Kocuria. The same or similar interpeptide bridges are present within Arthrobacter globiformis and its close relatives Arthrobacter pascens, Arthrobacter humi, and Arthrobacter oryzae as well as Arthrobacter crystallopoietes, Arthrobacter stackebrandtii, Arthrobacter methylotrophicus, Arthrobacter russicus, Arthrobacter ramosus, Arthrobacter monumenti, and Arthrobacter pigmenti. Because of the variability of quinone systems among Kocuria species, which includes MK-7(H2), MK-8(H2), MK-9(H2), or combinations of two of these components, this characteristic only distinguishes it from Arthrobacter species that possess completely unsaturated menaquinones.
Arthrobacter species with mono-saturated menaquinones can be unambiguously distinguished from Nesterenkonia because this genus contains only unsaturated menaquinones (MK-7, MK-8, or a combination of MK-7, MK-8, and MK-9). Differentiation of Arthrobacter species with completely unsaturated menaquinones from the majority of Nesterenkonia species can be achieved based on the length of the isoprenoic chain. While MK-8 and MK-9 are major components in certain Arthrobacter species, they are only reported for Nesterenkonia flava and Nesterenkonia alba (Luo et al., 2008; Luo et al., 2009). However, these two species have a peptidoglycan containing the interpeptide bridge Lys–Gly–Asp, which is not found in any Arthrobacter species. All Nesteronkonia species except Nesteronkonia lacusekhoensis, which contains a peptidoglycan with Lys–Glu in the interpeptide bridge, can be distinguished from Arthrobacter species because the former have an interpeptide bridge of Lys–Gly–Asp or Lys–Gly–Glu.
The peptidoglycan interpeptide bridge consisting of Lys–Gly–Ala (Kusser and Fiedler, 1983) and menaquinone MK-9 of Renibacterium distinguishes it from Arthrobacter. Also Rothia species can be reliably differentiated from Arthrobacter because MK-7 is the predominant menaquinone and Lys–Ala is the peptidoglycan interpeptide bridge, the latter only being found in Arthrobacter crystallopoietes.
It is harder to differentiate Arthrobacter from Zhihengliuella. Similar to several Arthrobacter species, Zhihengliuella possesses a peptidoglycan interpeptide bridge with Lys–Ala–Glu and a quinone system dominated by MK-10 and MK-9. The presence of the cell-wall sugar tyvelose in Zhihengliuella (Tang et al., 2009; Zhang et al., 2007) might be useful for differentiation, but cell-wall sugars have been analyzed only for a limited number of Arthrobacter species and the importance of this trait for differentiation has not been substantiated. The problem in differentiation might be related to the proposal of the genus Zhihengliuella. This genus was described based on phylogeny of the rRNA gene, but the branching from other taxa had only low bootstrap support (51%), and phenotypic traits distinguishing it from the Arthrobacter nicotianae group, which possesses similar or identical quinones and peptidoglycan structure, were provided without considering all the species of the group. For instance, the major fatty acids of Zhihengliuella are C15:0 anteiso and C15:0 iso. For the Arthrobacter nicotianae group, only C15:0 anteiso was reported as a trait in the latter study. In fact, a number of Arthrobacter nicotianae group species contain both C15:0 anteiso and C15:0 iso as abundant fatty acids, such as Arthrobacter uratoxydans, Arthrobacter protophormiae, and Arthrobacter sulfureus (Funke et al., 1996). Moreover, Zhihengliuella species possess high 16S rRNA gene sequence similarities with Arthrobacter species from different groups. For instance, the similarities with species of the rRNA cluster 4 are in the range 96.0–97.0%, which is indicative of an affiliation with this rRNA cluster. Hence, the re-evaluation of the taxonomic status of the genus Zhihengliuella is desirable. Perhaps, analysis of recA or some other housekeeping genes will resolve its taxonomic status more clearly. A possible outcome of these studies may be the transfer of Arthrobacter with Lys–Ala–Glu in the interpeptide bridge to the genus Zhihengliuella.
Taxonomic comments
As presently defined, the genus Arthrobacter comprises species with significant phenotypic variability in their composition of quinones, peptidoglycan, cell-wall sugars and, to a lesser degree, polar lipids. In many cases, groupings based on shared certain chemotaxonomic traits are well supported by comparative 16S rRNA gene sequence analyses, and most intra-group 16S rRNA gene sequence similarities are above 96.0%. These results suggest the genus Arthrobacter should be dissected into a number of genera.
Based on phylogeny, 16S rRNA gene sequence similarities, peptidoglycan compositions and/or quinone systems, the following genus-like groups can be defined: “Arthrobacter globiformis group” (Arthrobacter sensu stricto; corresponding to rRNA cluster 1), “Arthrobacter aurescens group” (corresponding to rRNA cluster 2), “Arthrobacter oxydans group” (corresponding to rRNA cluster 3), “Arthrobacter protophormiae group” (corresponding to rRNA cluster 4), “Arthrobacter sulfureus group” (corresponding to subclade I), “Arthrobacter agilis group” (corresponding to subclade II), “Arthrobacter citreus group” (corresponding to subclade III), “Arthrobacter psychrolactophilus group” (corresponding to subclade IV), “Arthrobacter pigmenti group”, “Arthrobacter albus/cumminsii group”, and “Sinomonas group” (corresponding to subclade V). Species assigned to the different groups and their major characteristics are listed in Tables 2-12, and their differential characteristics are given in Tables 13-23.
| A. globiformisd | A. pascensd | A. humicola | A. oryzae | A. crystallopoietesd | A. ramosus | A. methylotrophus | |
|---|---|---|---|---|---|---|---|
| Peptidoglycan type and interpeptide bridge | A3α, Lys–Ala3 | A3α, Lys–Ala2 | A3α, Lys–Ala>2e | A3α, Lys–Ala>2e | A3α, Lys–Ala | A3α, Lys–Ala4 | A3α, Lys–Ala2–4 |
| Quinone system | MK-9(H2) | MK-9(H2) | MK-9(H2) | MK-9(H2) | MK-9(H2) | MK-9(H2) | MK-9(H2), [MK-10(H2)]f |
| Cell-wall sugars | Gal, Glc | Gal, Glc | Gal, Rha | Gal, Glc | Gal, Glc | Gal, Man, Rha |
- a Abbreviations: Ala, alanine; Asp, aspartic acid; Glu, glutamic acid; Gly, glycine; Lys, lysine; Ser, serine; Thr, threonine; Gal, galactose; Glc, glucose; GlcN, glucosamine; Man, mannose; Rha, rhamnose; Rib, ribose; Xyl, xylose; MK-9(H2) designates a menaquinone with nine isoprenoic units in the side chain, of which one unit is saturated.
- b If not indicated otherwise data are taken from the original species description.
- c 98.4–99.5% 16S rRNA gene sequence similarities.
- d Data from Schleifer and Kandler (1972), Keddie and Cure (1978), Collins and Jones (1981).
- e Only quantitative amino acid composition of the peptidoglycan was reported (Kageyama et al., 2008). Peptidoglycan type is concluded from the amino acid composition.
- f Compounds shown in brackets were detected in the range 5–19%.
| A. aurescens | A. histidinolovorans | A. ilicis | A. nicotinovorans | A. nitroguajacolicus | A. ureafaciens | |
|---|---|---|---|---|---|---|
| Peptidoglycan type and interpeptide bridged | A3α, Lys–Ala–Thr–Ala | A3α, Lys–Ala–Thr–Ala | A3α, Lys–Ala–Thr–Ala | A3α, Lys–Ala–Thr–Ala | A3α, Lys–Ala–Thr–Ala | A3α, Lys–Ala–Thr–Ala |
| Quinone systeme | MK-9(H2) | MK-9(H2) | MK-9(H2) | MK-9(H2) | MK-9(H2), [MK-8(H2), MK-10(H2)]f | MK-9(H2) |
| Cell-wall sugarsg | Gal, (Man) | Gal, Glc | Gal, Man, Rha | Gal, (Man) | Gal, (Man) |
- a For abbreviations, see footnote to Table 2.
- b If not indicated otherwise, data are taken from the original species description.
- c 97.6–99.7% 16S rRNA gene sequence similarities.
- d Data are from Schleifer and Kandler (1972) except Arthrobacter nicotinovorans and Arthrobacter nitroguajaolicus.
- e Data are from Collins and Jones (1981) except Arthrobacter nicotinovorans and Arthrobacter nitroguajaolicus.
- f Reported to be present in minor amounts.
- g Sugars listed in parentheses are present only in minor amounts. Data are from Keddie and Cure (1978) except Arthrobacter nicotinovorans and Arthrobacter nitroguajaolicus.
| A. chlorophenolicus | A. defluvii | A. niigatensis | A. oxydansd | A. phenanthrenivorans | A. polychromogenes | A. scleromae | A. sulfonivorans | A. alkaliphilus | |
|---|---|---|---|---|---|---|---|---|---|
| Peptidoglycan type and interpeptide bridge | A3α, Lys–Ser–Thr–Ala | A3α, Lys–Ser–Thr–Ala | A3α, Lys–Ser–Thr–Alae | A3α, Lys–Ser–Thr–Ala | A3α, Lys–Ser–Thr–Alaf | A3α, Lys–Ser–Thr–Ala | A3α, Lys–Ser–Thr–Ala | A3α, Lys–Ser–Thr–Alae | |
| Quinone systemg | MK-9(H2), MK-9, [MK-8, MK-11]h | MK-9(H2), [MK-8(H2), MK-7(H2)]i | MK-9(H2), [MK-8(H2, H4)] | MK-9(H2) | MK-8, MK-9(H2) | MK-9(H2)f | MK-8(H2), [MK-10] | MK-9(H2),[MK-10(H2)] | MK-9(H2), [MK-10(H2)] |
| Cell-wall sugars | Gal, Glc, Rha | Gal, Glc | Gal, Glc |
- a For abbreviations, see footnote to Table 2.
- b If not indicated otherwise, data are taken from the original species description.
- c 97.3–99.7% 16S rRNA gene sequence similarities.
- d Data from Schleifer and Kandler (1972), Keddie and Cure (1978), Collins and Jones (1981).
- e Only the qualitative amino acid content of the peptidoglycan was reported (Ding et al., 2009). Peptidoglycan type is concluded from the amino acid composition.
- f Kodama et al. (1992).
- g Compounds shown in brackets were detected in the range 5–19%.
- h Ding et al. (2009).
- i Minor amounts of these two menaquinones were reported.
| A. ardleyensis | A. arilaitensis | A. bergerei | A. creatinolyticus | A. mysorens | A. nicotianae | A. protophormiae | A. rhombi | A. soli | A. uratoxydans | |
|---|---|---|---|---|---|---|---|---|---|---|
| Peptidoglycan type and interpeptide bridge | A4α, Lys–Ala–Glud | Lys–Ala–Glue | Lys–Ala–Glue | A4α, Lys–Ala–Glu | A4α, Lys–Ala–Gluf | A4α, Lys–Ala–Glu | A4α, Lys–Ala–Glu | A4α, Lys–Ala–Glu | A4α, Lys–Ala–Glu | |
| Quinone systemg | MK-8, MK-9 | MK-8, MK-9 | MK-8, MK-9, [MK-7]h | MK-8, MK-9, [MK-7]h | MK-8, (MK-7; MK-9)i | |||||
| Cell-wall sugars | Gal, Glc, Man, Rib | Glc, Man, Rib | Gal, Glc |
- a For abbreviations, see footnote to Table 2.
- b If not indicated otherwise, data are taken from the original species description.
- c 96.5–99.7% 16S rRNA gene sequence similarities.
- d Only the quantitative amino acid content of the peptidoglycan was reported (Chen et al., 2005a). Peptidoglycan type is concluded from the amino acid composition.
- e Only the qualitative amino acid content of the peptidoglycan was reported (Chen et al., 2005a). Peptidoglycan type is concluded from the amino acid composition.
- f Data are not from the type of the species but from patent strain ATCC 31021 (Stackebrandt et al., 1983b).
- g Compounds shown in brackets were detected in the range 5–19%.
- h Data are from Collins and Kroppenstedt (1983).
- i Quinones in parentheses indicate minor compounds (Bendinger et al., 1992).
| A. antarcticus | A. gangotriensis | A. kerguelensis | A. psychrophenolicus | A. sulfureus | |
|---|---|---|---|---|---|
| Peptidoglycan type and interpeptide bridge | A4α, Lys–Glu | A4α, Lys–Glu | A4α, Lys–Glu | A4α, Lys–Glu | A4α, Lys–Glu |
| Quinone systemd | MK-9, MK-8, MK-10e | MK-9, MK-10, [MK-8] | MK-9, MK-8, [MK-10] | MK-10 [MK-9] | MK-9 or MK-9, MK-10 [MK-8]f |
| Cell-wall sugars | Gal, Glc, Rha | Glc | Gal, Glc |
- a For abbreviations, see footnote to Table 2.
- b If not indicated otherwise, data are taken from the original species description.
- c 97.2–98.8% 16S rRNA gene sequence similarities.
- d Compounds shown in brackets were detected in the range 5–19%.
- e Only major menaquinones reported (Pindi et al., 2010).
- f Yamada et al. (1976) reported in addition to MK-9 minor amounts of MK-10 for Arthrobacter sulfureum AJ 1448 = IAM 1488 (formerly Brevibacterium sulfureum). Collins et al. (1979) reported in addition to MK-9 significant amounts of MK-7, MK-8, and MK-10 for the type strain Brevibacterium sulfureum c79 (= NCIB 10355). Collins and Kroppenstedt (1983) reported the major menaquinones MK-9 and MK-10 and significant amounts of MK-8 for “Brevibacterium sulfureum” NCIB 10355.
| A. agilis | A. flavus | A. parietis | A. subterraneus | A. tecti | A. tumbae | |
|---|---|---|---|---|---|---|
| Peptidoglycan type and interpeptide bridge | A3α, Lys–Thr–Ala3 | A3α, Lys–Thr–Ala3 | A3α, Lys–Thr–Ala2 | A3α, Lys–Thr–Ala3 | A3α, Lys–Thr–Ala3 | A3α, Lys–Thr–Ala3 |
| Quinone systemd | MK-9(H2), MK-8(H2)e, f | MK-9(H2) | MK-9(H2), MK-10(H2) | MK-9(H2), MK-10(H2) | MK-9(H2), MK-10(H2), [MK-11(H2), MK-9] | MK-9(H2), MK-10(H2), [MK-11(H2), MK-7(H2)] |
| Cell-wall sugarsg | GlcN | Gal, Glc, Rib | Gal | Gal, Glc, Rib | Gal, (Man) | Gal |
- a For abbreviations, see footnote to Table 2.
- b If not indicated otherwise, data are taken from the original species description.
- c 97.3–99.6% 16S rRNA gene sequence similarities.
- d Compounds shown in brackets were detected in the range 5–19%.
- e Present in minor amounts.
- f Recent re-analysis of the quinone system of the type strain of Arthrobacter agilis revealed a quinone system composed of 91% MK-9(H2), 8% MK-8(H2) and 1% MK-10(H2) (H.-J. Busse, unpublished results).
- g Sugars listed in parentheses are present only in minor amounts.
| A. citreusd | A. gandavensis | A. koreensis | A. luteolus | |
|---|---|---|---|---|
| Peptidoglycan type and interpeptide bridge | A3α, Lys–Thr–Ala2 | A3α, Lys–Thr–Ala2 | A3α, Lys–Thr–Ala2 | A3α, Lys–Thr–Ala2 |
| Quinone systeme | MK-9(H2), MK-8(H2) | MK-9(H2) | MK-8(H2), MK-9(H2) | MK-9(H2), MK-8(H2)f |
| Cell-wall sugars | Gal+ | Rha | Rhae |
- a For abbreviations, see footnote to Table 2.
- b If not indicated otherwise, data are taken from the original species description.
- c 97.6–98.9% 16S rRNA gene sequence similarities.
- d Data from Schleifer and Kandler (1972), Keddie and Cure (1978), and Collins and Kroppenstedt (1983).
- e Each menaquinone representing >20% of the total quinone content.
- f Data from Lee et al. (2003).
| A. psychrolactophilus | A. stackebrandtii | A. psychrochitiniphilus | A. alpinus | A. russicus | |
|---|---|---|---|---|---|
| Peptidoglycan type and interpeptide bridge | A3α, Lys–Thr–Ala3 | A3α, Lys–Ala2 | A3α, Lys–Thr–Ala1–2d | A3α, Lys–Thr–Ala3 | A3α, Lys–Ala2 |
| Quinone systeme | MK-9(H2) | MK-9(H2), [MK-10(H2)] | MK-9(H2) | MK-9(H2)f | MK-9(H2) |
| Cell-wall sugars | Gal, Rha |
- a For abbreviations, see footnote to Table 2.
- b If not indicated otherwise, data are taken from the original species description. including tentatively assigned species Arthrobacter russicus.
- c 97.3–98.4% 16S rRNA gene sequence similarities.
- d Only quantitative amino acid content of the peptidoglycan was reported (Chen et al., 2005a). Peptidoglycan type is concluded from the amino acid composition.
- e Compounds shown in parentheses were detected in the range 5–19%.
- f Presence of minor amounts of MK-8(H2) and MK-9(H2) were reported as well.
| A. castelli | A. monumenti | A. pigmenti | |
|---|---|---|---|
| Peptidoglycan type and interpeptide bridge | A3α, Lys–Ala–Ser–Ala3 | A3α, Lys–Ala4 | A3α, Lys–Ala4 |
| Quinone systemd | MK-9(H2), [MK-10(H2)] | MK-9(H2) | MK-9(H2), [MK-7(H2), MK-10(H2)] |
| Cell-wall sugarse | Gal, (Xyl, Rha) | Gal, (Xyl, Rha) | Gal, Rha |
- a For abbreviations, see footnote to Table 2.
- b If not indicated otherwise, data are taken from the original species description.
- c 96.2–97.9% 16S rRNA gene sequence similarities.
- d Compounds shown in brackets were detected in the range 5–19%.
- e Sugars listed in parentheses are present only in minor amounts.
| A. albus | A. cumminsii | |
|---|---|---|
| Peptidoglycan type and interpeptide bridgeb | A4α, Lys–Ala–Glu | A4α, Lys–Ser(Gly)–Glu |
| Quinone system | MK-8(H2) | MK-8(H2) |
- a For abbreviations, see footnote to Table 2.
- b 99.1% 16S rRNA gene sequence similarity.
| S. atrocyanea | S. flava | A. albidus | A. echigonensis | |
|---|---|---|---|---|
| Peptidoglycan type and interpeptide bridge | A3α, Lys–Ser–Ala2–3 | A3α, Lys, Gly, Ala4c | A3αd, Lys, Ser, Ala | A3αd, Lys, Ser, Ala |
| Quinone systeme | MK-9(H2), [MK-8(H2)] | MK-9(H2), [MK-8(H2)] | MK-9(H2), [MK-(102)] | MK-9(H2) [MK-10(H2)] |
| Cell-wall sugars | Gal, Glc, Man |
- a For abbreviations, see footnote to Table 2.
- b 96.0–99.3% 16S rRNA gene sequence similarities.
- c Only the amino acid content of the peptidoglycan was reported (Zhou et al., 2009). Peptidoglycan type and number of alanines are concluded from the analysis of relative amounts of amino acids in the peptidoglycan.
- d Only the amino acid content of the peptidoglycan was reported (Ding et al., 2009). Peptidoglycan type is concluded from the amino acids detected in the peptidoglycan.
- e Compounds shown in brackets were detected in the range 5–19%.
| Characteristic | A. globiformisb | A. humicolab | A. oryzaeb | A. pascensb | A. crystallopoietes | A. methylotrophusc | A. ramosus |
|---|---|---|---|---|---|---|---|
| Major fatty acidsd | C15:0 anteiso, [C15:0 iso], (C16:0 iso, C17:0 anteiso) | C15:0 anteiso, (C17:0 anteiso, C16:0 iso, C15:0 iso) | C15:0 anteiso, [C17:0 anteiso] | C15:0 anteiso, [C17:0 anteiso], (C15:0 iso) | C15:0 anteiso, (C17:0 anteiso) | C15:0 anteiso, (C15:0 iso, C17:0 anteiso, C16:0 iso) | C15:0 anteiso, (C17:0 anteiso, C15:0 iso) |
| Motility | − | + | + | − | −e | − | +e |
| NaCl range for growth (%, w/v) | 0–5 | 0–3 | 0–2 | 0–5 | nr | 0–2.5 | nr |
| Utilization of L-arabinose | + | − | − | + | nr | nr | nr |
| Nitrate reductase | − | − | + | − | +f | − | nr |
| Pyrrolidonyl arylamidase | − | − | + | − | nr | nr | nr |
| Urease | − | − | − | − | +f | nr | nr |
| Enzyme assay (API ZYM): | |||||||
| Esterase lipase (C8) | − | w | − | − | nr | nr | nr |
| Acid phosphatase | − | + | + | − | nr | nr | nr |
| α-Galactosidase | − | + | − | + | nr | nr | − |
| β-Glucuronidase | − | w | + | − | nr | nr | nr |
| α-Glucosidase | + | + | + | + | nr | nr | − |
| α-Mannosidase | + | w | − | + | nr | nr | − |
- a Symbols and abbreviations: +, positive; w, weakly positive; −, negative; nr, not reported. Data are from the original descriptions of the species if not indicated otherwise.
- b Kageyama et al. (2008).
- c Borodina et al. (2002b).
- d Fatty acids in brackets are found in the range between >10–25%; fatty acids in parentheses are present in the range 5–10%. Data from Funke et al. (1996), Borodina et al. (2002b), and Kageyama et al. (2008).
- e Keddie et al. (1986).
- f Li et al. (2004c).
| Characteristic | A. aurescens | A. histidinolovorans | A. ilicis | A. nicotinovorans | A. nitroguajacolicus | A. ureafaciens |
|---|---|---|---|---|---|---|
| Major fatty acidsc | C15:0 anteiso, (C15:0 iso) | C15:0 anteiso, [C17:0 anteiso], (C16:0 iso) | C15:0 anteiso, (C16:0 iso, C15:0 iso, C14:0 iso) | C15:0 anteiso, [C17:0 anteiso] | C15:0 anteiso, (C15:0 iso, C16:0 iso, C17:0 anteiso) | C15:0 anteiso, C17:0 anteiso, (C15:0 iso, C16:0 iso) |
| Oxidase | + | nr | −d | nr | + | nr |
| Hydrolysis of: | ||||||
| Esculin | + | nr | − | nr | + | nr |
| Starch | + | − | − | v | + | − |
| Elastase | + | nr | − | nr | + | nr |
| Pyrrolidonyl arylamidase | − | nr | + | nr | − | nr |
| Motility | − | − | + | nr | + | − |
| Utilization of: | ||||||
| Gluconate | + | nr | − | nr | − | nr |
| L-Histidine | + | + | + | + | nr | − |
| L-Leucine | − | − | − | v | nr | − |
| L-Rhamnose | − | w | w | + | nr | − |
| Inositol | nr | + | − | + | nr | + |
| Assimilation of: | ||||||
| D-Xylose | w | + | + | nr | − | + |
| Uridine | − | nr | + | nr | − | nr |
| Arbutin | w | nr | − | nr | w | nr |
| Sucrose | − | nr | + | nr | + | nr |
| Propionic acid | − | + | w | + | + | + |
| Cyclodextrin | + | nr | − | nr | − | nr |
| D-Melibiose | w | nr | w | nr | − | nr |
| 3-Methylglucose | w | nr | w | nr | − | nr |
| D-Raffinose | w | nr | w | nr | + | nr |
| Salicin | − | nr | w | nr | − | nr |
| Urea formed from creatine | nr | + | − | + | nr | + |
- a Symbols and abbreviations: +, positive; −, negative; w, weakly positive reaction; v, variable. Data are from the original description of the species if not indicated otherwise.
- b Data from Kotoucková et al. (2004) and Kodama et al. (1992).
- c Fatty acids in brackets are found in the range beween >10–25%; fatty acids in parentheses are present in the range 5–10%.
- d Positive in Collins et al. (1981).
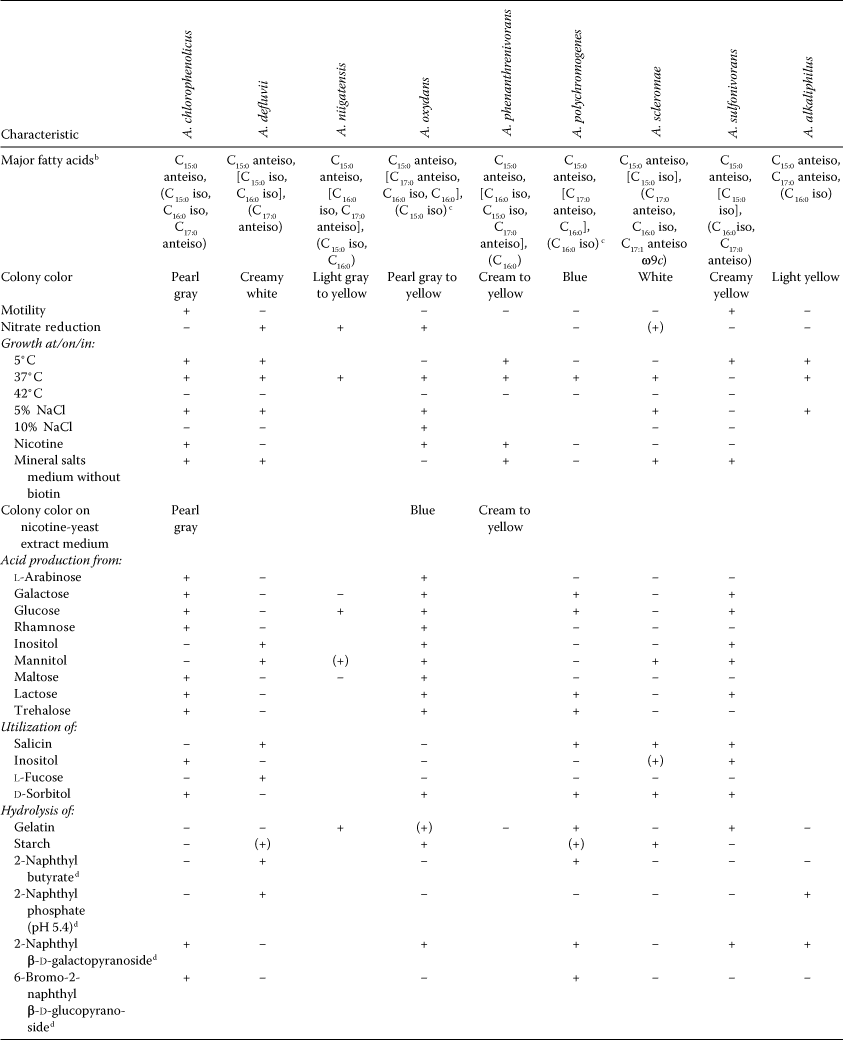
- a Symbols and abbreviations: +, positive; (+), weakly positive; −, negative. Data are from the original descriptions of the species and from Kim et al. (2008) if not indicated otherwise.
- b Fatty acids in brackets are found in the range beween >10–25%; fatty acids in parentheses are present in the range 5–10%.
- c Data from Kodama et al. (1992).
- d Data from API ZYM tests. The intensity of the color was measured on a scale from 0 to 5 and was interpreted as negative at values of 0 or 1 and positive at values of 2–5 (Mudarris et al., 1994).
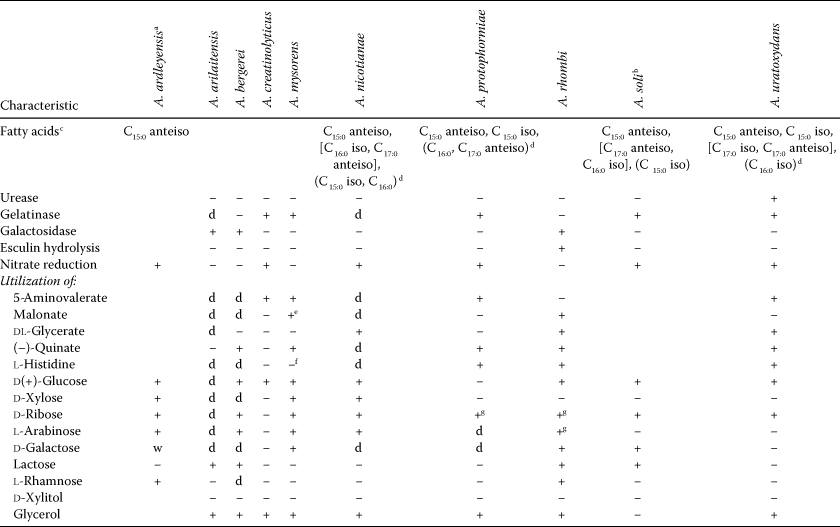
- a Physiological data were generated using the Biolog test.
- b Physiological data were generated using API 20NE and API 50CH test strips (Roh et al., 2008b).
- c Fatty acids representing >10–25% of the total are shown in squared brackets, fatty acids representing 5–10% of the total are given in parentheses.
- d Data published by Funke et al. (1996)
- e Negative in Stackebrandt et al. (1983b).
- f Positive in Stackebrandt et al. (1983b).
- g Negative in Osorio et al. (1999).
| Characteristic | A. gangotriensisa | A. kerguelensisa | A. psychrophenolicusb | A. sulfureusf, b |
|---|---|---|---|---|
| Fatty acidsc | C15:0 anteiso, [C15:0 iso, C16:0 iso,], (C17:0 anteiso, C17:0 iso, C18:1) 4–30 | C15:0 anteiso, C17:0 anteiso, (C15:0 iso, C17:0 iso) | C15:0 anteiso, (C15:0 iso, C16:0 iso, C17:0 anteiso) | C15:0 anteiso, [C15:0 iso], (C16:0 iso, C17:0 anteiso)d |
| Temperature range for growth (°C) | 4–30 | |||
| Optimum growth temperature (°C) | 22 | 22 | 25 | 25–30 |
| Nitrate reduction | + | − | ||
| Esculin hydrolysis | − | + | − | |
| Casein hydrolysis | + | + | − | |
| Skimmed milk hydrolysis at 10°C | + | − | ||
| Lysine decarboxylase | − | + | − | |
| Acid production from: | ||||
| D-Galactose | + | − | + | |
| D-Mannose | + | − | − | |
| Sucrose | − | − | − | + or − |
| D-Xylose | − | + | − | − |
| Utilization of sole carbon sources: | ||||
| D-Cellobiose | + | − | + | |
| D-Mannitol | − | − | + | |
| D-Rhamnose | − | + | + | |
| Trehalose | − | + | + | |
| L-glutamic acid | − | + | + | |
| L-Histidine | − | + | + | |
| L-Tryptophan | − | − | + | |
| Assimilation of: | ||||
| Glucose | − | + | ||
| Phenylacetate | + | − | ||
| DNA G+C content (mol%) | 66 | 58 | 64.5–66 |
| Characteristic | A. agilisb | A. flavusb | A. parietisb | A. subterraneusc | A. tectib | A. tumbaeb |
|---|---|---|---|---|---|---|
| Major fatty acidsd | C15:0 anteiso, [C15:0 iso], (C16:1 iso) | C15:0 anteiso, [C16:0 iso, C17:0], (C17:0 anteiso) | C15:0 anteiso, C15:0 iso, (C17:0 anteiso) | C15:0 anteiso, [C15:0 iso, C17:1 iso, C17:0 anteiso], (C16:0 iso) | C15:0 anteiso, C15:0 iso, (C17:0 anteiso) | C15:0 anteiso, [C15:0 iso, C17:0 anteiso] |
| Alkaliphilic | − | − | pH 5.3–10.5 | − | + | |
| Growth at 5% NaCl | − | + | + | + | + | + |
| Oxidase | + | − | − | − | − | − |
| Nitrate reduction | − | − | + | − | − | v |
| β-Glucuronidase | − | − | − | + | ||
| N-Acetyl-β-glu-cosaminidase | − | − | + | − | ||
| Urease | − | − | v | − | − | v |
| Gelatin hydrolysis | + | + | + | (+) | (+) | |
| Alkaline phosphatase | − | − | +w | (−)w | ||
| Esterase lipase (C8) | − | + | (+) | (+)w | ||
| Lipase (C14) | − | + | − | − | − | − |
| α-Galactosidase | + | − | − | (−) |
- a Symbols and abbreviations: +, all strains positive; (+), ≥80 % positive; v, 21–79% positive; (−), ≤20% positive; −, all strains negative; w, all positive reactions are weak.
- b Data from Heyrman et al. (2005).
- c Data from Chang et al. (2007).
- d Fatty acids in brackets are found in the range beween >10–25%; fatty acids in parentheses are present in the range 5–10%.
| Characteristic | A. citreusb | A. gandavensisb | A. koreensisc | A. luteolusb |
|---|---|---|---|---|
| Major fatty acidsd | C15:0 anteiso, [C16:0], (C15:0 iso, C16:0 iso, C17:0 anteiso) | C15:0 anteisoe, C15:0 iso and C17:0 anteisof | C15:0 anteiso, C15:0 iso | C15:0 anteiso, [C15:0 iso] |
| β-Galactosidase | + | + | − | − |
| Urease | − | + | nr | − |
| Hydrolysis of: | ||||
| Esculin | − | + | nr | − |
| L-Isoleucine-AMC | + | − | nr | + |
| p-Nitrophenyl phosphate | − | + | nr | − |
| p-Nitrophenyl β-D-glucoside | − | + | nr | − |
| Proline-and leucine-p-nitroanilide | − | + | − | |
| p-Nitrophenyl phosphate | − | + | nr | − |
| ONPG and p-nitrophenyl α-D-galactoside | − | + | nr | − |
| Utilization of: | ||||
| D-Arabitol | nr | nr | + | − |
| p-Hydroxyphenylacetic | nr | nr | + | − |
| Glycerol | nr | nr | − | + |
| Tween 80 | + | nr | + | − |
| L-Arabinose | + | nr | − | − |
- a Symbols and abbreviations: +, positive; −, negative; AMC, 7-Amino-4-methylcoumarin; ONPG, ortho-nitro-phenyl-β-D-galactopyranoside; nr, not reported.
- b Data from Storms et al. (2003).
- c Data from Lee et al. (2003).
- d Fatty acids in brackets are found in the range beween >10–25%; fatty acids in parentheses are present in the range 5–10%.
- e Predominant.
- f Significant.
| Characteristic | A. alpinus | A. psychrochitiniphilus | A. psychrolactophilus | A. stackebrandtii | A. russicus |
|---|---|---|---|---|---|
| Major fatty acidsc | C15:0 anteiso, [C16:0 iso, C17:0 anteiso], (C15:0 iso) | C15:0 anteiso, [C17:0 anteiso] | C15:0 anteiso, [C17:0 anteiso], (C16:0 iso) | C15:0 anteiso, [C15:0 iso, C16:0 iso, C17:0 anteiso] | C17:0 anteiso, [C15:0 anteiso,] |
| Growth at 30°C | − | − | wd | +† | + |
| Nitrate reduction | − | + | −d | +d | − |
| Hydrolysis of urea | + | − | −d | +d | − |
| Chitinase | nr | + | − | nr | nr |
| Gelatinase | nr | − | + | nr | nr |
| Enzyme assays (API ZYM): | |||||
| α-Glucosidase | + | nr | +d | + | nr |
| β-Galactosidase | + | nr | +d | wd | nr |
| β-Glucuronidase | + | nr | wd | + | nr |
| Assimilation/utilization of: | |||||
| L-Arabinose | + | nr | −d | +d | + |
| D-Maltose | + | nr | +d | +d | − |
| Lactose | + | + | +† | −d | − |
| Tween 40 | w | nr | −d | +d | + |
| Tween 80 | − | + | −d | −d | + |
| Reaction in the Biolog test: | |||||
| Raffinose | nr | − | + | nr | − |
| Cyclodextrin | nr | nr | − | + | − |
| Tween 40 | nr | + | − | + | + |
| N-Acetyl-D-mannosamine | nr | nr | + | − | − |
| L-Arabinose | nr | + | − | + | − |
| L-Fucose | nr | nr | − | + | nr |
| D-Melezitose | nr | nr | + | − | − |
| Methyl-D-galactoside | nr | + | − | + | − |
| Methyl-D-glucoside | nr | nr | + | − | − |
| Stachyose | nr | nr | − | + | − |
| L-Asparagine | nr | + | − | + | − |
| Uridine | nr | + | − | + | v |
- a Symbols and abbreviations: +, positive; w, weakly positive; −, negative; v, variable; nr, not reported.
- b Data are from the original descriptions of the species if not indicated otherwise.
- c Fatty acids in brackets are found in the range beween >10–25%; fatty acids in parentheses are present in the range 5–10%.
- d Data from Zhang et al. (2010).
| Characteristic | A. castelli | A. monumenti | A. pigmenti |
|---|---|---|---|
| Major fatty acidsc | C15:0 anteiso, [C15:0 iso, C16:0 iso], (C17:0 anteiso) | C15:0 anteiso, C15:0 iso | C15:0 iso, C15:0 anteiso, (C17:0 anteiso) |
| Alkaliphilic | − | − | + |
| Nitrate reduction | − | + | v |
| Urease | + | v | − |
| Gelatin hydrolysis | − | + | + |
| Esterase lipase (C8) | + | v | w |
| Lipase (C14) | w | − | − |
- a Symbols and abbreviations: +, all strains positive; v, variable; −, all strains negative; w, weak reaction
- b Data from Heyrman et al. (2005).
- c Fatty acids in brackets are found in the range beween >10–25%; fatty acids in parentheses are present in the range 5–10%.
| Characteristic | S. atrocyaneab | S. flavab | A. albidusc | A. echigonensisc |
|---|---|---|---|---|
| Major fatty acidsd | C15:0 anteiso, C17:0 anteiso, [C15:0 iso,,], (C16:0 iso, C17:0 iso) | C15:0 anteiso, C15:0 iso, [C17:0 anteiso] | C15:0 anteiso, C17:0 anteiso, [C15:0 iso], (C16:0 iso) | C15:0 anteiso, [C15:0 iso, C17:0 anteiso], (C16:0 iso) |
| Optimum pH | 6–8 | nr | 7 | 7 |
| Optimum temperature (°C) | 30–37 | nr | 25–37 | 25–37 |
| Reduction of nitrate | + | + | − | − |
| Pyrrolidonyl arylamidase | nr | nr | + | + |
| Urease | + | + | + | − |
| Hydrolysis of gelatin | − | − | − | − |
| Utilization of: | ||||
| Maltose | + | nr | − | (+) |
| Mannitol | + | + | + | − |
| Ribose | + | nr | + | + |
| Sucrose | + | + | (+) | + |
| Xylose | nr | nr | (+) | − |
| Esterase (C4) | nr | nr | (+) | + |
| Cystine arylamidase | nr | nr | + | + |
| Chymotrypsin | nr | nr | − | − |
| Galactosidase | nr | nr | − | − |
| β-Glucosidase | + | + | + | − |
New groups proposed for the genus Arthrobacter
“Arthrobacter globiformis group” (Arthrobacter sensu stricto; rRNA cluster 1)
Members of the Arthrobacter “globiformis group” (Table 2) exhibit 98.4–99.5% 16S rRNA gene similarity, a peptidoglycan type A3α (Lys–Ala2 or Lys–Ala3) and menaquinone MK-9(H2). Arthrobacter crystallopoietes is preliminarily assigned to this group because it shares highest 16S rRNA gene sequence similarity (97.6%) with Arthrobacter globiformis and possesses a peptidoglycan composed of Lys–Ala. Table 13 lists differential characteristics for this group.
“Arthrobacter aurescens group” (rRNA cluster 2)
Species of this group (Table 4) share high 16S rRNA similarities (>97.5%), peptidoglycan type A3α (Lys–Ala–Thr–Ala) and the menaquinone MK-9(H2). Table 14 lists differential characteristics for this group.
“Arthrobacter oxydans group” (rRNA cluster 3)
Close relatedness of the species of this group is indicated by high 16S rRNA gene similarities (>97%). The peptidoglycan type is A3α (Lys–Ser–Thr–Ala). For most species, MK-9(H2) is the major menaquinone, but MK-8(H2) predominates in Arthrobacter scleromae. In Arthrobacter phenanthrenivorans both MK-8 and MK-9(H2) are abundant (Table 4). Table 15 lists differential characteristics for this group.
“Arthrobacter protophormiae group” (rRNA cluster 4)
The species of this group (Table 5) possess 16S rRNA gene similarities of 95.5–99.7%. Each species shares at least 96.7% similarity with another species within this group, but most values are higher than 97.5%. Members of this group share the peptidoglycan type A4α (Lys–Ala–Glu) and a quinone system composed of completely unsaturated menaquinones with eight and/or nine isoprenoic units and with MK-8 always predominant. Arthrobacter rhombi is assigned to this group based on peptidoglycan type. Table 16 lists differential characteristics for this group.
“Arthrobacter sulfureus group” (subclade I)
The species in this group (Table 6) share at least 97.2% 16S rRNA gene similarity and form a robust clade supported by high bootstrap values in phylogenetic trees. Members of this group share the peptidoglycan type A4α (Lys–Glu) and a quinone system composed of completely unsaturated menaquinones, with MK-9 predominating but also usually significant amounts of MK-10. Table 17 lists differential characteristics for this group.
“Arthrobacter agilis group” (subclade II)
Species comprising this group (Table 7) share 97.3–99.6% similarity in the 16S rRNA gene and form a subclade supported by high bootstrap values. The peptidoglycan type is A3α with Lys–Thr–Ala2–3. MK-9(H2) is the predominant menaquinone, but significant amounts of MK-10(H2) are present in four species of the subclade. Table 18 lists differential characteristics for this group.
“Arthrobacter citreus group” (subclade III)
Species comprising this group (Table 8) share 16S rRNA gene similarities of 97.6–98.9% and form a subclade supported by moderate bootstrap values. The peptidoglycan type is A3α with Lys–Thr–Ala2. The quinone system comprises menaquinone MK-9(H2) or almost equal amounts of MK-8(H2) and MK-9(H2). Table 19 lists differential characteristics for this group.
“Arthrobacter psychrolactophilus group” (subclade IV)
Species comprising this group (Table 9) share >97.0% similarity in the 16S rRNA gene and form a robust clade supported by 99% bootstrap values. The interpeptide bridge of the peptidoglycan is either Lys–Thr–Ala1–3 or Lys–Ala2, and MK-9(H2) is the predominant menaquinone. Moderate proportions of MK-10(H2) may be present as well. Species of this group are psychrophilic or psychrotolerant and grow at 0–4°C. It is interesting that in a FASTA sequence comparisons among 28 database entries showing 97.8% or higher similarity with Arthrobacter psychrochitiniphilus, 26 sequences were from Antarctica, Spitzbergen, glaciers, or other sources likely to permit low temperature growth (Bai et al., 2006; Cheng and Foght, 2007; Hansen et al., 2007; Reddy et al., 2009; Srinivas et al., 2009). Hence, growth at low temperatures may be another trait of the members of this group. Table 20 lists differential characteristics for this group.
“Arthrobacter pigmenti group
The core of this group (Table 10) is represented by Arthrobacter pigmenti and Arthrobacter castelli, which share 97.9% 16S rRNA gene similarity. Arthrobacter monumenti is assigned to this group (96.2% similarity) although it shares higher 16S rRNA gene similarities (but still less than 97.0%) with some other Arthrobacter species. However, chemotaxonomic traits of Arthrobacter monumenti are in better agreement with those of Arthrobacter pigmenti and Arthrobacter castelli. Arthrobacter monumenti shares with Arthrobacter pigmenti and Arthrobacter castelli a peptidoglycan with four alanines in the interpeptide chain (Lys–Ala4 or Lys–Ala–Ser–Ala3) and the cell-wall sugars Gal and Rha. Notably, the same interpeptide bridge is found in some species of the “Arthrobacter globiformis group”, including Arthrobacter ramosus and Arthrobacter methylotrophus. Thus, future studies might combine the “Arthrobacter globiformis group” and “Arthrobacter pigmenti group” or transfer species between the groups. Table 21 lists differential characteristics for this group.
“Arthrobacter albus/cumminsii group”
So far, only two species (Table 11) are assigned to this group. They share 99.1% similarity in the 16S rRNA gene and higher sequence similarities with representatives of other genera, such as Kocuria, Citricoccus, and Micrococcus, than with other Arthrobacter species. The peptidoglycan type is A4α with Lys–Ala–Glu or Lys–Ser–(Gly)–Glu. The quinone system is MK-8(H2). Table 22 lists differential characteristics for this group.
“Sinomonas group” (subclade V)
The two species Arthrobacter echinogenes and Arthrobacter albidus (Table 12) form a clade with the two species of the genus Sinomonas (Zhou et al., 2009). The type strains of the two species share more than 97.0% 16S rRNA gene similarity with at least one of the two species of the genus Sinomonas and less than 94.5% similarity with other recognized Arthrobacter species. Arthrobacter albidus and Arthrobacter echigonensis possess Lys, Ser, and Ala in their peptidoglycan, which is consistent with the peptidoglycan type A3α (Lys–Ser–Ala2–3) reported for Sinomonas atrocyanea. The menaquinone is MK-9(H2). For these reasons, Arthrobacter echinogenes and Arthrobacter albidus should be transferred to the genus Sinomonas. Table 23 lists differential characteristics for this group.
Species not assigned to the above groups
Due to discrepancies between the 16S rRNA gene sequence and phenotypic data, several species remain ungrouped in this scheme, including Arthrobacter alkaliphilus, Arthrobacter crystallopoietes, Arthrobacter methylotrophus, Arthrobacter nasiphocae, Arthrobacter ramosus, Arthrobacter roseus, Arthrobacter russicus, Arthrobacter sanguinis, and Arthrobacter woluwensis. However, if priority is given to the chemotaxonomic characteristics, several species can be tentatively assigned to certain of the above-mentioned “Arthrobacter groups”.
Arthrobacter alkaliphilus and Arthrobacter methylotrophicus form a separate line of descent within the radiation of the genus Arthrobacter. While the high 16S rRNA gene similarity of 97.4% suggest a close relationship, similarities with other species are not much lower, 97.1% to Arthrobacter nicotinovorans and 96.9% to Arthrobacter histidinolovorans. The peptidoglycan structure of Arthrobacter alkaliphilus (Lys–Ser–Thr–Ala) and the quinone system of MK-9(H2) with significant amounts of MK-10(H2) distinguishes it from Arthrobacter methylotrophicus, which contains Lys–Ala2–4 and MK-9(H2) but no MK-10(H2). Arthrobacter nicotinovorans and Arthrobacter histidinolovorans are also distinguishable from Arthrobacter alkaliphilus based on peptidoglycan structure and quinone system. The peptidoglycan structure of Arthrobacter alkaliphilus is characteristically that of “Arthrobacter oxydans group”, and hence Arthrobacter alkaliphilus is tentatively placed in this group.
Phylogenetic analyses of the rRNA genes of Arthrobacter crystallopoietes do not yield a stable assignment to any phylogenetic subclade. It shares 97.6% 16S rRNA gene similarity with the type species of the genus, Arthrobacter globiformis, and no other species possesses higher similarity. Sequence similarity values with other species of “Arthrobacter globiformis group” (Arthrobacter sensu stricto; rRNA cluster 1) are in the range of 96.2–97.1%. The peptidoglycan interpeptide bridge consists of Lys–Ala, which is similar to that of other rRNA cluster 1 species. The cell-wall sugars Gal and Glc are also found in other species of this group. Hence, Arthrobacter cyrystallopoietes is tentatively placed in “Arthrobacter globiformis group”.
Arthrobacter nasiphocea possesses low 16S rRNA gene similarity, 95.5–96.5%, with Arthrobacter woluwensis, Arthrobacter pascens, Citricoccus alkalitolerans, Arthrobacter oryzae, Arthrobacter methylotrophus, Arthrobacter nitroguajacolicus; Arthrobacter phenanthrenivorans, Arthrobacter roseus, Arthrobacter humicola, Arthrobacter oxydans, and Arthrobacter polychromogenes. Likewise, phylogenetic analyses do not suggest a clear affiliation with any of the Arthrobacter groups. The unrelatedness of Arthrobacter nasiphocea to other Arthrobacter species is also indicated from the peptidoglycan structure (Lys–Ala2-Gly2–3–Ala [Gly]) and quinone system MK-9(H2) + MK-8(H2). Hence, Arthrobacter nasiphocea might be considered a representative of a novel genus. Table 24 lists differential phenotypic characteristics for this species relative to some other ungrouped species.
| A. nasiphocea | A. roseus | A. sanguinis | A. woluwensis | |
|---|---|---|---|---|
| Major fatty acidsa | nr | C14:0 iso, C14:0, C15:0 iso, C15:0 anteiso, C15:0, C16:0 iso, C16:0, C16:1, C17:0 anteiso, C18:0, and C18:2b | C15:0 anteiso and C17:0 anteisoc | C15:0 anteiso, [C15:0 iso, C16:0 iso, C17:0 anteiso], (C17:0 iso) |
| Colony color | Grayish-white | Red | Whitish-grayish | Whitish-grayish |
| Peptidoglycan type | Lys–Ala2–Gly2–3–Ala (Gly) (A3α) | Lys–Gly–Ala3 (A3α) | nr | L–Lys–D-Asp (A4α) |
| Quinone system | MK-9(H2), MK-8(H2) | MK-9(H2) | nr | nr |
| Growth at 37°C | + | − | nr | + |
| Growth at 5°C | − | + | nr | nr |
| Hydrolysis of: | ||||
| Esculin | − | − | nr | + |
| Tween 80 | nr | nr | nr | nr |
| Urease | − | − | − | + |
| Nitrate reduction | − | + | − | − |
| Utilization of: | ||||
| Inositol | nr | − | − | − |
| Sorbitol | nr | + | + | + |
| Lactose | nr | − | − | + |
| Xylitol | nr | nr | − | − |
| Xylose | nr | + | − | − |
| Enzyme activity (API ZYM): | ||||
| Ester lipase C8 | − | nr | + | + |
| Lipase C14 | − | nr | − | + |
| Cystine arylamidase | − | nr | − | + |
| Trypsin | − | nr | + | + |
| β-Glucosidase | − | nr | − | + |
| α-Fucosidase | − | nr | − | − |
| α-Galactosidase | − | nr | + | − |
| β-Galactosidase | − | − | + | + |
| α-Mannosidase | − | nr | + | + |
| N-Acetyl-β-glucosaminidase | − | nr | + | + |
- a Fatty acids in brackets are found in the range beween >10–25%; values in parentheses are present in the range 5–10%; nr, not reported.
- b Only qualitative values were provided by Reddy et al. (2002).
- c Predominant.
In both maximum likelihood and neighbor-joining phylogenetic analyses of the 16S rRNA gene, Arthrobacter methylotrophus is associated with Arthrobacter alcaliphilus but without bootstrap support. The 16S rRNA gene sequences of these two species share 97.4% similarity, but similarities with certain species of “Arthrobacter globiformis group”, “Arthrobacter aurescens group”, “Arthrobacter oxydans group”, and “Arthrobacter agilis group” are not much lower, 96.9–97.3%. Hence, 16S rRNA data do not unambiguously indicate an affiliation of Arthrobacter methylotrophus to any specific Arthrobacter group. It contains the menaquinone MK-9(H2), which is widely distributed in arthrobacters and not helpful for classification of this species. The peptidoglycan interpeptide bridge is Ala2–4 and also similar to those found in other groups, including representatives of the “Arthrobacter globiformis group”, “Arthrobacter pigmenti group”, and “Arthrobacter psychrolactophilus group”. Since 16S rRNA gene sequence similarities with species of the latter two groups are significantly lower, Arthrobacter methylotrophus is tentatively assigned to the “Arthrobacter globiformis group”.
Arthrobacter ramosus exhibits 16S rRNA gene similarities of 97.2–98.2% with species of the “Arthrobacter aurescens group”, but phylogenetic analyses do not provide support for this or any other affiliation (Figure 1). The peptidoglycan interpeptide bridge of Lys–Ala4 is also not consistent with an association with the “Arthrobacter aurescens group”, whose members possess Lys–Ala–Thr–Ala. Sequence similarities with species of “Arthrobacter globiformis group” and Arthrobacter methylotrophicus are in the range of 96.3–96.8% and 96.3–97.1%, respectively, which also does not support a clear affiliation to these groups. Nevertheless, based on moderate sequence similarities and a similar peptidoglycan interpeptide bridge, Arthrobacter ramosus is tentatively placed in the “Arthrobacter globiformis group”.
In some phylogenetic trees, Arthrobacter roseus is placed at the root of the “Arthrobacter psychrolactophilus group” but without bootstrap support for assignment to this group. High 16S rRNA gene similarities of 97.0–97.6% identify the “Arthrobacter oxydans group” as the next closest relatives. However, the composition of its peptidoglycan interpeptide bridge, Lys–Gly–Ala3, is unique within Arthrobacter and does not provide clear evidence for an assignment to any of the designated Arthrobacter groups. Likewise, the menaquinone is MK-9(H2), which is common within the genus. Hence, this species may be a representative of a novel genus within the Micrococcaceae. Table 24 lists differential phenotypic characteristics for this species relative to some other ungrouped species.
Phylogenetic analyses suggest that Arthrobacter russicus is closely related to Renibacterium salmoninarum, a relationship that is also supported by a high 16S rRNA gene similarity of 97.2%. However, these species differ significantly in G+C content (65.5 and 56.3 mol%, respectively; Li et al., 2004c; Wiens et al., 2008); in the quinone system, and in peptidoglycan structure. Arthrobacter russicus has menaquinone MK-9(H2) and a peptidoglycan with Lys–Ala2, whereas Renibacterium salmoninarum has MK-9 and peptidoglycan with Lys–Ala–Gly (Kusser and Fiedler, 1983; Sanders and Fryer, 1980). These differences may be explained by the evolution of Renibacterium salmoninarum from an Arthrobacter-like ancestor, largely via genome reduction (Wiens et al., 2008), adaption to the salmonid host, and loss of some biosynthetic capabilities. Nevertheless, the absence of significant phenotypic similarities does not support the reclassification of Arthrobacter russicus within the genus Renibacterium. Compared to Arthrobacter globiformis, the 16S rRNA gene of Arthrobacter russicus possesses a 12-bp insertion (position 421–432), which is also found in Renibacterium salmoninarum and other moderately related Arthrobacter species, such as Arthrobacter psychrolactophilus (95.5% 16S rRNA gene sequence similarity), Arthrobacter stackebrandtii (95.4% 16S rRNA gene sequence similarity), and Arthrobacter agilis (94.9% 16S rRNA gene sequence similarity). In other species, such as Arthrobacter pigmenti, Arthrobacter castelli, Arthrobacter albus, and Arthrobacter cumminsii, the insertion at this position is a few nucleotides shorter. Whereas peptidoglycan structures clearly distinguish Arthrobacter russicus from Arthrobacter psychrolactophilus and Arthrobacter agilis, no differences in this trait were reported for Arthrobacter russicus and Arthrobacter stackebrandtii. In sequence comparisons where this insertion is removed from the 16S rRNA gene sequence, Arthrobacter globiformis is among the species sharing highest similarities (>96.0%). Since Arthrobacter russicus branches in phylogenetic trees which do not include Renibacterium salmoninarum at the root of the “Arthrobacter psychrolactophilus group” (Ding et al., 2009; Pindi et al., 2010; Tvrzová et al., 2005b; Zhang et al., 2010) and shares with Arthrobacter stackebrandtii the peptidoglycan type (Lys–Ala2) and the quinone system MK-9(H2), this species is tentatively placed in “Arthrobacter psychrolactophilus group”.
Arthrobacter sanguinis shares 16S rRNA gene similarities of 94.0–95.2% with Arthrobacter species from different Arthrobacter groups, such as Arthrobacter crystallopoietes, Arthrobacter cumminsii, Arthrobacter globiformis, Arthrobacter phenanthrenivorans, Arthrobacter pascens, Arthrobacter albus, and Arthrobacter bergerei but also with the type strain of Kocuria rosea. Phylogenetically, Arthrobacter sanguinis forms a separate line of descent with Acaricomes phytoseiuli, but this clade is not supported by high bootstrap values, and the two species share only 93.9% 16S rRNA gene similarity. Although it is obvious from these data that Arthrobacter sanguinis is not a member of Arthrobacter sensu stricto or any of the other Arthrobacter groups described herein, it is not placed in a new group because important information is not available, including both the quinone system and peptidoglycan structure. If chemotaxonomic data were available, it might support classification with Acaricomes phytoseiuli in the genus Acaricomes or placement in a novel genus. Table 24 lists differential phenotypic characteristics for this species relative to some other ungrouped species.
Among species of the genus Arthrobacter, Arthrobacter woluwensis is the only species with Asp in the peptidoglycan interpeptide bridge, which is designated peptidoglycan type A4α (Schleifer and Kandler, 1972) or A11.31 (http://www.peptidoglycan-types.info). Other Arthrobacter species with an A4α type interpeptide bridge possess Glu alone (A11.33), Ala–Glu (A11.35), or Ala–Ser–Glu (A11.38, A11.48, or A11.58). Sequence similarities of 96.7–97.6% in the 16S rRNA gene suggest that this species is a relative of the Arthrobacter globiformis group, but there is no bootstrap support for this assignment. Three species of the Arthrobacter nicotianae group, two species of Arthrobacter oxydans group, and Arthrobacter methylotrophus, Arthrobacter crystallopoietes, Arthrobacter roseus, and Arthrobacter alkaliphilus also possess similarity values within this range or slightly lower, 96.4–96.8%. In the absence of a clear assignment to a subclade one rRNA cluster, the unique peptidoglycan of Arthrobacter woluwensis suggests that it represents a novel genus, and hence it is not assigned to any of the other Arthrobacter groups. Table 24 lists differential phenotypic characteristics for this species relative to some other ungrouped species.
Degradation potential
Numerous arthrobacters have been examined for their potential to degrade xenobiotics and other harmful substances.
Three Arthrobacter strains isolated from soil surrounding an outdoor coal storage pile grew with naphthalene vapor as sole source of carbon. However, none of the known genes for naphthalene degradation were detected, such as nahAc, nahAd, nahH, phnAC, xzlE, or GST (Dore et al., 2003). Several phenanthrene-degrading Arthrobacter strains have been isolated after long-term exposure to the polycyclic aromatic hydrocarbons (Bodour et al., 2003).
Arthrobacter aurescens TC1 metabolizes 23 different s-triazine compounds including atrazine, which it utilizes as a sole source of carbon and nitrogen. Catabolic enzymes involved are TrzN, AtzB, and AtzC (Sajjaphan et al., 2004; Strong et al., 2002). The amidohydrolase TrzN is a zinc-containing enzyme that has a broader specificity than AtzA, the corresponding enzyme of Pseudomonas sp. strain ADP and displaces cyano, azido, halide, S-alkyl, and O-alkyl substituents (Shapir et al., 2006; Strong et al., 2002). Further studies suggest that the genes trzN, atzB, and atzC are located on a 380-kb plasmid (Sajjaphan et al., 2004). Another atrazine degrader, Arthrobacter AD1 is very closely related to Arthrobacter ureafaciens on the basis of 16S rRNA gene similarity. Its atzA gene is located on the chromosome (Cai et al., 2003). Three atrazine-degrading strains closely related to Arthrobacter crystallopoietes were isolated from French soil (Rousseaux et al., 2001), and the catabolic genes were located on a 117-kb plasmid (Rousseaux et al., 2002).
Degradation of 4-chlorophenol has been reported for several arthrobacters. Arthrobacter ureafaciens CPR706 first eliminates the chloro-substituent to form hydroquinone. This strain also degrades other para-substituted phenols, including 4-nitro-, 4-bromo-, 4-iodo-, and 4-fluorophenol via the hydroquinone pathway. It does not degrade ortho- or meta-substituted phenols (Bae et al., 1996). The type strain of Arthrobacter defluvii and a second strain of this species were isolated by Kim et al. (2008) after stimulation of a sewage sample with 4-chlorophenol. Another Arthrobacter strain, described as Arthrobacter chlorophenolicus (Westerberg et al., 2000) was isolated after successively increasing the 4-chlorophenol concentration in a soil suspension from 50 to 350 p.p.m. over a period of 165 d. Two strains of Arthrobacter nitroguajacolicus were enriched from forest soil and shown to degrade 4-nitroguaiacol. A third strain of this species produces 4-nitroguaiacol in the course of 4-nitrophenol transformation (Kotoučková et al., 2004).
Arthrobacter sp. strain SU DSM 20407 hydrolytically dehalogenates 4-chlorobenzoate, a breakdown product of polychlorinated biphenyls. The degradation requires three genes organized in an operon on the plasmid pASU1. These genes, designated fchA, fchB, and fchC, encode 4-chlorobenzoate:CoA ligase, 4-chlorobenzoyl-CoA dehalogenase, and 4-hydroxybenzoyl-CoA thioesterase (Schmitz et al., 1992; Zhuang et al., 2003). The same set of genes is also present in Arthrobacter sp. strain TM-1, which was originally isolated from sewage sludge by enrichment on 4-chlorobenzoate. Arthrobacter sp. strain TM-1 dehalogenates 4-chlorobenzoate, and the product, 4-hydroxybenzoate, is further metabolized via protocatechuate. However, other chlorinated benzoates as well as benzoate itself do not support growth and are presumably not metabolized (Marks et al., 1984). The first enzyme in the pathway, 4-chlorobenzoate:CoA ligase produces 4-chlorobenzoyl CoA and AMP (Zhou et al., 2004). The second enzyme is a trimeric protein, 4-chlorobenzoyl CoA dehalogenase, and has a secondary and three-dimensional structure similar to that of the corresponding enzyme from Pseudomonas sp. CBS-3. Dehalogenase activity in Arthrobacter sp. strain TM-1 is also inhibited by dissolved oxygen and stimulated by manganese (Zhou et al., 2008b).
Cullington and Walker (1999) isolated a bacterial strain D47 capable of degrading the substituted phenylurea herbicides diuron, isoproturon, chlorotoluron, linuron, monolinuron, and monuron. Turnbull et al. (2001) showed that this strain is closely related to Arthrobacter oxydans and Arthrobacter polychromogenes and that it hydrolyzes the urea side chain at the carbonyl group. Arthrobacter RC100 contains three plasmids, and two of them are involved in mineralization of carbaryl (1-naphthyl N-methylcarbamate), which is one of the most commonly used carbamate pesticides for the control of a wide variety of insects. During the mineralization, metabolites that appear early in the suspension are 1-naphthol and salicylic acid. Arthrobacter sp. RC100 grows also on salicylic acid, 1-naphthol, gentisic acid, protocatechuic acid, salicylaldehyde, and 2-naphthol (Hayatsu et al., 1999).
The N-heteroaromatic quinaldine (2-methylquinoline) is utilized by Arthrobacter nitroguajacolicus Rü61a (formerly assigned to the species Arthrobacter ilicis) as a source of carbon, nitrogen, and energy. The degradation proceeds via the “anthranilate pathway” (Parschat et al., 2003). The genes for conversion of quinaldine to anthranilate are located on a the conjugative linear catabolic plasmid pAL1, which comprises 112,992 bp (Parschat et al., 2007).
Arthrobacter protophormiae RKJ100 utilizes o-nitrobenzoate as sole source of carbon, nitrogen, and energy via an oxygen insensitive pathway that releases ammonia to the culture medium. The catabolic genes are located on a 65-kb plasmid (Chauhan and Jain, 2000). The strain degrades o-nitrobenzoate via a reductive pathway leading to the formation of o-hydroxylaminobenzoate and anthranilate (Pandey et al., 2003). Also p-nitrophenol and 4-nitrocatechol are degraded by Arthrobacter protophormiae RKJ100, and the corresponding genes are encoded from the same 65-kb plasmid (Chauhan et al., 2000).
“Arthrobacter keyseri” 12B (originally designated Micrococcus strain 12B) possesses high 16S rRNA gene similarity to Arthrobacter ureafaciens, but the name has never been validly published and hence it does not have standing in nomenclature. The strain converts phthalate to protocatechuate through cis-3,4-dihydroxy-3,4-dihydrophthalate and 3,4-dihydroxyphthalate. The genes of the catabolic pathway are plasmid encoded, and sequence analysis reveals genes encoding a transposon resolvase, enzymes required for catabolism of protocatechuate (3,4-dihydroxybenzoate), a putative ATP-binding cassette transporter, a possible phthalate ester hydrolase, a fragment of a norfloxacin resistance-like transporter, and enzymes required for the conversion of phthalate to protocatechuate, respectively (Eaton, 2001).
Arthrobacter sp. strain JBH1 was isolated after enrichment from a nitroglycerin-contaminated site, and it can grow with nitroglycerin as sole source of carbon, nitrogen, and energy. The strain shares 99.5% 16S rRNA gene similarity with Arthrobacter pascens, indicating that it is a representative of the genus Arthrobacter sensu stricto. It converts nitroglycerin via 1,2-dinitroglycerin to glycerin, which serves as an energy source (Husserl et al., 2010).
Arthrobacter nicotinovorans has the ability to use the tobacco plant alkaloid nicotine as its sole source of carbon and energy. The catabolic genes are located on the 165-kb plasmid pAO1, which also confers the ability to take up nicotine from the medium (Baitsch et al., 2001; Igloi and Brandsch, 2003). Furthermore, the plasmid encodes enzymes essential for biosynthesis of the molybdenum dinucleotide cofactor of nicotine dehydrogenase as well as open reading frames necessary for the uptake and utilization of carbohydrates, sarcosine, and amino acids. pAO1 was successfully transferred by conjugation to a pAO1-negative strain, so genes for conjugation must be present as well.
Enzymes
While a number of enzymes have been identified and characterized in arthrobacters, many are from strains whose identifications are ambiguous. Many of these strains are assigned to the genus Arthrobacter on the basis of rod–coccus morphological cycle, presence of lysine in the peptidoglycan, fatty acid profile, and/or 16S rRNA analyses. Since these phenotypic traits are not restricted to arthrobacters and the 16S rRNA gene analysis is sometimes misleading, it is likely that in the future some of the Arthrobacter strains listed below will be assigned to other genera.
A strain of Arthrobacter ilicis associated with the marine sponge Spirastrella sp. produces an extracellular serine type acetylcholinesterase with maximum activity at 45°C and pH 8.0 (Mohapatra and Bapuji, 1998). The enzyme activity is unaffected by Mg2+ and Ca2+ but inhibited by higher concentrations (25 mM) Co2+, Cu2+, Hg2+, Mn2+, Ni2+ and Zn2+ as well as EDTA.
Arthrobacter FR-3, isolated from sediment samples of a swamp near the Dow's Lake, Ottawa, Canada, produces a serine-type sulfide oxidase. The free and immobilized sulfide oxidase shows optimal activity at pH 7.5 and 6.0, respectively, and at 35°C. Enzyme activity is not inhibited by 1 mM Ca2+, Mg2+, Na+, and EDTA, but it is completely inhibited by 1 mM Co2+ and Zn2+ (Mohapatra et al., 2006).
The psychrophilic Arthrobacter sp. strain TAD20 produces two cold-adapted, heat labile chitinases, ArChiA and ArChiB. The chitinases are active at 4°C but not at 17 or 24°C. They lose 50–60% activity after incubation at 50°C for 30 min. Deduced from the gene sequences, the primary structures of ArChiA and ArChiB consist of 846 and 539 amino acids, respectively. Metals are not required for activity, and their activity is unaffected by 10 mM EDTA (Lonhienne et al., 2001).
Three different β-galactosidases have been detected in Arthrobacter B7. The sizes of the isozymes are 111 kDa, 71 kDa, and 52 kDa. The 111-kDa isozyme exhibits 22% amino acid sequence similarity with the EbgA protein of Escherichia coli, but the optimum temperature is approximately 20°C lower than that of the Escherichia coli β-galactosidase. The enzyme requires Mn2+ or Mg2+ at 1 mM for maximum activity; Ca2+ supports 61% activity, whereas Cu2+ is completely inhibitory (Trimbur et al., 1994). The amino acid sequence of the 71-kDa isozyme (designated group lacG) has little similarity with the enzymes of the lacZ family. Highest similarity was found with β-galactosidases from endospore-formers, such as the thermophile Geobacillus stearothermophilus (21.8%) and Bacillus circulans (17.7%). The enzyme is active over a pH range from 6 to 9 and activity is optimum at pH 6.8. The enzyme activity is reduced after treatment with EDTA but can be restored by addition of 5 mM Mg2+, 10 mM Mn2+, or Ca2+; 10 mM Co2+ also increases the activity whereas Ni2+ has no effect and Cu2+ is inhibitory. The activity is optimal at 45–50°C and is quite stable for at least 70 h at 25, 30, and 35°C (Gutshall et al., 1995). The 52-kDa β-galactosidase of Arthrobacter B7 is active as a dimer and hydrolyzes β-1,4 or β-1,3 glycoside linkages. The amino acid sequence is most similar to human and mouse acid β-galactosidase and a corresponding enzyme of Xanthomonas manihotis (Gutshall et al., 1997).
The psychrophile Arthrobacter strain SB from the Antarctic is a close relative of Arthrobacter sulfonivorans. The strain produces a β-galactosidase (BgaS) which exhibits optimal activity at 18°C, 50% activity at 0°C, but is inactivated within 10 min at 37°C. At 4°C, the active enzyme is a homotetramer but dissociates into inactive monomers at 25°C. After dissociation into monomers, the enzyme does not regain activity even when cooled to 4°C. The size of the monomer is approximately 116 kDa. The enzyme is active from pH 6.0 to 9.5, with greatest activity at pH 7.0. Activity is dependent on Mg2+, whereas other monoor divalent cations have only small stimulatory effects. EDTA inactivates the enzyme at a concentration of 100 mM within 90 min. The nucleotide sequence of the gene that encodes BgaS has highest similarity with those of lacZ-like genes of Arthrobacter sp. C2–2 and Arthrobacter psychrolactophilus (Coker et al., 2003). A cold-adapted β-galactosidase was also characterized from another Antarctic strain, Arthrobacter sp. 32c (Hildebrandt et al., 2009). This strain is a close relative of Arthrobacter oxidans and Arthrobacter polychromogenes. The gene sequence of the enzyme has highest similarity with those of Arthrobacter FB24 (77.1%) and Arthrobacter aurescens TC1 (71.8%). The enzyme has a temperature optimum at 50°C, with residual activity being 60% at 25°C, and 15% at 0°C. The pH optimum is 6.5, but more than 90% activity remains at pH 8.5. Treatment with 100 mM EDTA for 2 h does not affect the activity. Metal ions such as Na+, K+, Mg2+, Co2+, or Ca2+ do not affect activity, whereas Mn2+, Fe2+, and Ni2+ strongly and Cu2+ or Zn2+ completely inhibit the enzyme.
Arthrobacter D10, which was isolated from soil in Pennsylvania, produces an extracellular alkaline phosphatase (de Prada et al., 1996). At pH 9.1, the activity is maximal at 45–50°C. Further examination demonstrated two extracellular alkaline phosphatases, designated D10A and D10B. Alkaline phosphatase D10B requires calcium for optimal activity, whereas D10A has equal activity with or without calcium. D10A has a maximum pH around 9.5 and is active in the pH range 7.0–11.0. D10B has a narrower pH range, with a maximum at pH 9.0 and less than 10% activity at pH 7.0 and 9.5. D10A loses 60% of activity within 15 min at 55°C, and the less heat-labile D10B loses 20% of activity within 15 min at 55°C.
The soil isolate Arthrobacter strain KM possesses activities for several inorganic polyphosphate [poly(P)]- and ATP-dependent kinases including glucokinase, NAD kinase, mannokinase, and fructokinase (Mukai et al., 2003). An enzyme purified from this strain phosphorylates glucose and mannose utilizing either poly(P) or ATP and was designated poly(P)/ATP-glucomannokinase. The enzyme is a monomer with a molecular mass of approximately 30 kDa. The corresponding gene consists of 804 bp. The enzyme has highest activity at pH 7.5 and 45°C, but 50% activity is lost after treatment at 40°C for 5 min. Both enzyme functions, poly(P)- and ATP-dependent gluco- and mannokinase activities require Mg2+ or Mn2+. Zn2+ and Co2+ mediate high activity only for poly(P)-dependent glucomannokinase activity (Mukai et al., 2003).
A pyridoxal-5′-phosphate-dependent D-threonine aldolase has been purified and characterized from Arthrobacter sp. DK-38 (Kataoka et al., 1997). The 51-kDa enzyme catalyzes cleavage of D-threonine into glycine and acetaldehyde and the reverse reaction forming D-threonine and D-allothreonine. It is inhibited by chelating reagents such as EDTA, and activity is dependent on divalent cationic salts, including MnCl2, MgCl2, CoCl2, NiCl2, and to a lesser degree FeSO4, whereas CaCl2 has little effect. Other metallic salts including CuSO4, ZnSO4, NaCl, KCl, AlCl2, and FeCl2 have no effect, and Ag2SO4, HgCl2, and CdCl2 completely inhibit the enzyme activity. MnCl2 also contributes significantly to thermal stability of the enzyme (Kataoka et al., 1997).
Two extracellular proteinases and an extracellular proline iminopeptidase have been characterized from Arthrobacter nicotianae 9458, isolated from the bacterial smear of surface ripened cheeses (Smacchi et al., 1999a, 1999b). The proteinases have molecular masses of approximately 54 kDa (proteinase P1) and 71 kDa (proteinase P2). Both proteinases degrade αs1-and β-casein rapidly at pH 6.5 and 37°C. The optima of the two enzymes are pH 9.5 and 9.0, respectively, but both are still active at pH 6.5. At their optimal pH, the temperature optima are 37°C and 55–60°C, respectively, and both enzymes remain active at 15°C. P1 is irreversibly inactivated after exposure to 70°C for 1 min. The second proteinase, P2, retains 50% of its activity under the same conditions but is inactivated at 85°C after 1 min. EDTA has a strong inhibitory effect on P1 but is only moderately inhibitory to P2. Both enzymes are inhibited by 1 mM Zn2+, Hg2+, Ni2+, Cu2+, or Co2+. At higher concentrations, Fe2+ and Mn2+ are inhibitory as well. The proline iminopeptidase has a molecular mass of about 54 kDa. Its temperature optimum is 37°C, but activity decreases rapidly above 42°C. Its pH optimum is at 8.0, but 60% of the maximal activity is retained at pH 9.0. It is completely inhibited after treatment for 1 min at 80°C. EDTA is moderately inhibitory. Fe2+ and Sn2+ completely inactivate the enzyme, and only a little activity is observed in the presence of Zn2+, Hg2+, Ni2+, Cu2+, or Co2+. Mn2+ is moderately inhibitory, and Ca2+ and Mg2+ have little effect.
Genes for degradation of creatinine to glycine and formaldehyde have been identified in Arthrobacter sp. TE1826. Together with the repressor gene soxR, the soxA, crnA, and creA genes are organized in an operon. The crnA gene encodes a 258-amino acid protein, and the creA gene encodes a 441-amino acid protein (Nishiya et al., 1998).
A gene encoding a dimethylglycine dehydrogenase (dmg) has been identified in Arthrobacter globiformis NRRL-B2979, and soxBDAG genes encoding the subunits of the heterotetrameric sarcosine oxidase have been identified in Arthrobacter spp. 1-IN (Meskys et al., 2001). The dmg gene encodes a protein of 830 amino acids whose molecular mass is approximately 90 kDa.
Enrichment and isolation procedures
Due to the high number of species within the genus and their different physiological properties, no general procedure for enrichment and isolation can be recommended. The majority of Arthrobacter species are unexacting and grow on standard media containing peptone and yeast extract at or near neutral pH, such as nutrient broth or trypticase soy broth. As shown by Funke et al. (1996), who examined the antibiotic sensitivity of 24 Arthrobacter strains, resistance to ciprofloxacin, clindamycin, and gentamicin is widespread among this genus. Hence, supplementation of the enrichment and isolation medium with these antibiotics may be suitable.
Maintenance procedures
Standard procedures for conservation are recommended, such as storage of culture in 25% glycerol at −80°C or lyophilization.
List of species of the genus Arthrobacter
Arthrobacter globiformis
(Conn 1928) Conn and Dimmick 1947, 301AL (Basonym: Bacterium globiforme Conn 1928, 3.)
glo.bi.for'mis. L. n. globus ball, globe; L. adj. suff. -formis -is -e (from L. n. forma figure, shape, appearance) -like, in the shape of; N.L. masc. adj. globiformis spherical.
Nonmotile. Colonies on yeast extract-peptone media show no distinctive pigmentation. Either nutritionally nonexacting or require biotin alone. Growth occurs in a suitable mineral salts medium with an ammonium salt or nitrate as sole nitrogen source (with biotin if required) and with glucose as carbon + energy source. The widely used Conn strain ATCC 4336 (NCIB 8602) requires biotin; the type strain does not. Starch is hydrolyzed, nicotine is not utilized. The cell-wall peptidoglycan is of the Lys–Ala3 type, A3α (Fiedler et al., 1970). The whole-cell sugars are galactose and glucose. The principal isoprenoid quinone is MK-9(H2) (Collins et al., 1979). The predominant fatty acid is C15:0 anteiso (48%), followed by C15:0 iso (21%), C17:0 anteiso (7%), C16:0 iso (7%), C16:0 (4%), C14:0 iso (2%), and C17:0 iso (2%). All other fatty acids are at levels below 1% (Funke et al., 1996).
Source: soil.
DNA G+C content (mol%): 62.0–65.5 (Tm).
Type strain: AS 1.1894, ATCC 8010, BCRC (formerly CCRC) 10598, CCUG 581, CCUG 12157, CCUG 28997, CIP 81.84, DSM 20124, HAMBI 88, HAMBI 1863, IAM 12438, ICPB 3434, NBRC 12137, JCM 1332, LMG 3813, NCIMB 8907, NRIC 0151, NRRL B-2979, VKM Ac-1112.
Sequence accession no. (16S rRNA gene): X80736.
Sequence accession no. (recA): AF214780
Additional remarks: after continuously decreasing oxygen tension in the growth medium, the type strain of Arthrobacter globiformis is able to grow in the presence of nitrate, glucose and pyruvate as carbon sources carrying out anaerobic respiratory nitrate reduction with fermentation of carbon sources and also to grow anaerobically in the absence of nitrate, with glucose and pyruvic acid as carbon sources (Eschbach et al., 2003).
Arthrobacter agilis
(Ali-Cohen 1889) Koch, Schumann and Stackebrandt 1995, 837VP (Micrococcus agilis Ali-Cohen 1889, 36)
a'gi.lis. L. masc. adj. agilis agile.
Spheres (0.8–1.2 µm in diameter) occur in pairs and tetrads. Agar colonies are circular, entire, slightly convex, smooth, and matte. Sediment is formed in nutrient broth. No growth occurs on Simmons' citrate medium. Good growth and dark rose-red pigmentation occur on agar slants. The pigment is water insoluble. Motile by means of one to three flagella. Nonmotile strains may occur. Spores are not formed. Gram-stain-positive. The cell-wall peptidoglycan type is type L-Lys–L-Thr–L-Ala (variation A3α). The predominant menaquinone isoprenologue is MK-9(H2); MK-8(H2) is a minor component. Contains major amounts of anteiso methyl-branched acids (~65.5% C15:0 anteiso) and smaller amounts of iso methyl-branched acids (~13% C15:0 iso and ~6% C16:1 iso). The polar lipids are phosphatidylglycerol, diphosphatidylglycerol, phosphatidylinositol, and an unknown ninhydrin-negative glycolipid. The amino sugar in the cell-wall polysaccharide is glucosamine. Chemo-organotrophic. Metabolism is respiratory. Aerobic. Acid and gas are not produced from glucose, mannose, lactose, galactose, and glycerol. Catalase-positive. Porphyrin respiratory enzymes are produced. Oxidase-positive. Acetylmethylcarbinol is not produced. β-Galactosidase (o-nitrophenyl-β-D-galactopyranoside test) positive. Methyl red negative. Indole and hydrogen sulfide are not produced. Nitrate is not reduced. Gelatin, starch, and esculin are hydrolyzed. Arginine dihydrolase, ornithine and lysine decarboxylases, and phenylalanine deaminase are not produced. Tween 80 may be split. DNase may be produced. Urease, tyrosinase, and phosphatase are not produced. Beta-hemolysis does not occur. Good growth occurs at temperatures between 20 and 30°C. No growth occurs at 37°C. No growth occurs on medium containing 5% NaCl. Susceptible to penicillin, streptomycin, chloramphenicol, tetracycline, erythromycin, novobiocin, ampicillin, carbenicillin, and gentamicin. Resistant to lysozyme. Saprophytic.
Source: water, soil, and human skin.
DNA G+C content (mol%): 67.0–69.0 (Tm).
Type strain: ATCC 966, DSM 20550, CCM 2390, CIP 81.67, NBRC 15319, JCM 2584, LMG 17244, VKM B-1973.
Sequence accession no. (16S rRNA gene): X80748.
Sequence accession no. (recA): AF214779
Arthrobacter albidus
Ding, Hirose and Yokota 2009, 860VP
al'bi.dus. L. masc. adj. albidus whitish, referring to the color of the colonies.
Cells are nonmotile and non-spore-forming. Gram-stain positive, catalase-positive, oxidase-negative. Shows a rod–coccus growth cycle and produces non-fluorescent pigment. Growth occurs on nutrient broth agar at 10–40°C, optimal temperature for growth is 30°C. Grows in the presence of 3–7% (w/v) NaCl, but no growth occurs in the presence of 7% NaCl. The pH range for growth is 6–10, and the optimum pH is 7. Colonies are round, convex, glossy, with entire margins and are white or light yellow. Using the API CORYNE system, positive reactions are observed for activities of pyrazinamidase, pyrrolidonyl arylamidase, and urease and for the utilization of ribose and mannitol. Negative reactions are obtained for nitrate reduction, hydrolysis of gelatin, and for the utilization of maltose, lactose, and glycogen. Utilization of glucose, xylose, and sucrose is weak. Using the API ZYM system, activity is detected for alkaline phosphatase, esterase lipase C8, leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, naphthol-AS-BI-phosphohydrolase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, and α-mannosidase. No activity is detected for lipase C14, achymotrypsin, α-galactosidase, N-acetyl-β-glucosaminidase, or α-fucosidase. Esterase C4 and acid phosphatase activities are weak. The predominant fatty acids are C15:0 anteiso, C17:0 anteiso, and C15:0 iso. The diagnostic diamino acid of the cell-wall peptidoglycan is lysine and the major components are lysine, serine, and alanine. The menaquinone composition is MK-9(H2) (83%), MK-10(H2) (14%), and MK-8(H4) (2%).
Source: a filtration substrate made from volcanic rock from Niigata, Japan.
DNA G+C content (mol%): 70.8 (HPLC).
Type strain: LC13, CCTCC 206018, IAM 15386, JCM 21830.
Sequence accession no. (16S rRNA gene): AB248533.
Arthrobacter albus
Wauters, Charlier, Janssens and Delmée 2000b, 1699VP (Effective publication: Wauters, Charlier, Janssens and Delmée 2000a, 2415.)
al'bus. L. masc. adj. albus white, because of the white colonies of the organism.
Cells are small coryneform bacteria, nonmotile and without spore formation. Colonies are white and are 1 mm in diameter after 48 h of incubation at 37°C on blood agar. Metabolism is obligately aerobic. Catalase is positive. Urease and esculin are not hydrolyzed, and nitrates are not reduced. Gelatin is slowly and weakly liquefied, and there is no DNase activity. Tyrosine is not hydrolyzed. Simmons' citrate is negative. Pyrrolidonyl peptidase is positive. The organism is resistant to desferrioxamine. No acid is produced from carbohydrates, and there is no utilization of these substrates in the API 50 CH system. Alkaline and acid phosphatase, phosphoamidase, esterase, esterase-lipase, leucine arylamidase, and trypsin are present but not lipase, valine arylamidase, cystine arylamidase, chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetylglucosaminidase, α-mannosidase, and α-fucosidase. The main cellular fatty acid is C15:0 anteiso, and the peptidoglycan type is L-Lys–L-Ala–L-Glu.
Source: human clinical specimens.
DNA G+C content (mol%): not determined.
Type strain: CF-43, DSM 13068, ATCC BAA-273, CCUG 43812, CIP 106791, JCM 11943.
Sequence accession no. (16S rRNA gene): AJ243421.
Arthrobacter alkaliphilus
Ding, Hirose and Yokota 2009, 859VP
al.ka.li'phi.lus. N.L. n. alkali (from the Arabic word al-qaliy) the ashes of saltwort; Gr. adj. philos loving; N.L. masc. adj. alkaliphilus loving alkaline environments.
Cells are nonmotile and non-spore-forming. Gram-stain positive, catalase-positive, and oxidase-negative. Shows a rod–coccus growth cycle and produces non-fluorescent pigment. Growth occurs on nutrient broth agar at 5–40°C, optimal temperature for growth is 30°C. Grows in the presence of 3–7% (w/v) NaCl. The pH range for growth is 6–11 and the optimum pH is 8.5. Colonies are round, convex, glossy, with entire margins and are light yellow. Using the API CORYNE system, positive for pyrazinamidase, pyrrolidonyl arylamidase, and urease activity and for glucose, ribose, maltose, and mannitol utilization. Negative reactions are obtained for nitrate reduction, for hydrolysis of gelatin, and for utilization of glycogen, sucrose, and xylose. Lactose gives a weak reaction. Using the API ZYM system, activity is detected for alkaline phosphatase, acid phosphatase, esterase lipase C8, leucine arylamidase, valine arylamidase, trypsin, naphthol-AS-BI-phosphohydrolase, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, and α-mannosidase. No activity is detected for esterase C4, lipase C14, cystine arylamidase, β-glucosidase, N-acetyl-β-glucosaminidase, or α-fucosidase. The activity of α-chymotrypsin is weak. Predominant fatty acids are C15:0 anteiso, C17:0 anteiso, and C16:0 iso. The diamino acid of the cell-wall peptidoglycan is lysine and the major components are lysine, serine, threonine, and alanine. The menaquinone composition is MK-9(H2) (77%), MK-10(H2) (17%), and MK-8(H4) (3%).
Source: a filtration substrate made from volcanic rock from Niigata, Japan.
DNA G+C content (mol%): 69.0 (HPLC).
Type strain: LC6, CCTCC AB 206013, IAM 15383, JCM 21827.
Sequence accession no. (16S rRNA gene): AB248527.
Arthrobacter alpinus
Zhang, Schumann, Liu, Xin, Zhou, Schinner and Margesin 2010, 2152VP
al.pi'nus. L. masc. adj. alpinus alpine inhabitant, referring to the isolation of this strain from an alpine environment.
Cells are irregular rods showing a rod–coccus cycle, 0.8–1.0 × 1.3–1.9 µm after 3 d at 20°C on R2A agar plates, often occurring in pairs as typical V-forms. Cells are Gram-stain-positive, aerobic, and nonmotile. Colonies on R2A agar and on nutrient agar are creamy to yellow 176 after 3 d and yellow after 6 d, round, convex, shiny, and with an entire margin; colony diameter is 1–1.5 mm after 3 d at 20°C on R2A agar. Growth occurs in liquid R2A medium and on agar plates at 1–25°C, with fastest growth rates at 20–25°C; growth is absent at 30°C. On R2A agar plates, growth occurs at pH 6–9 and in the presence of 0–5% (w/v) NaCl. Produces catalase but not cytochrome oxidase. Negative for nitrate reduction, indole production, H2S production, and citrate utilization. Esculin, urea, and starch are hydrolyzed but not casein and Tween 80. Negative for activities of arginine dihydrolase, lysine dihydrolase, ornithine dihydrolase, lipase (C14), N-acetyl-β-glucosaminidase, trypsin, α-chymotrypsin, α-fucosidase, and pyrrolidonyl arylamidase. Positive for activities of alkaline phosphatase, acidic phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, naphthol-AS-BI-phosphohydrolase, α-glucosidase, β-glucosidase, α-galactosidase, β-galactosidase, β-glucuronidase, and α-mannosidase. Utilizes D-glucose, lactose, L-arabinose, D-maltose, D-mannose, D-mannitol, and N-acetylglucosamine but not citrate, capric acid, adipic acid, and phenylacetic acid as sole carbon source; utilization of Tween 40 is weak. Negative for fermentation of glucose, ribose, xylose, mannitol, maltose, lactose, sucrose, glycogen, inositol, sorbitol, rhamnose, melibiose, amygdalin, and L-arabinose. Sensitive to ampicillin, penicillin G, streptomycin, tetracycline, and chloramphenicol (each 30 µg/ml), but resistant to cyclosporin A (100 µg/ml). The predominant cellular fatty acids of the type strain are C15:0 anteiso, C16:0 iso, and C17:0 anteiso. Contains MK-9(H2) as the major menaquinone and minor amounts of MK-8(H2) and MK-10(H2). The cell-wall peptidoglycan is of the type A3α L-Lys–L-Thr–Ala3. The predominant cell-wall sugars are galactose and rhamnose.
Source (type strain): an alpine soil in Fuschertörl, Hohe Tauern, Austria.
DNA G+C content (mol%): 61.9 (Tm).
Type strain: S6-3; DSM 22274, CGMCC 1.8950.
Sequence accession no. (16S rRNA gene): GQ227413.
Arthrobacter antarcticus
Pindi, Manorama, Begum and Shivaji 2010, 2265VP
an.tarc'ti.cus. L. masc. adj. antarcticus southern, used to refer to the Antarctic, referring to the place from where the organism was isolated.
Cells have rod–coccus cycle. Rod-shaped cells grown in tryptone soya broth (TSB) at 22°C for 72 h are 1.5–2.2 µm long and 0.2–0.3 µm wide. Cells are Gram-stain-positive, motile, aerobic, and form a yellow colony on TSB agar plates. Growth occurs between 4–25°C, but not at 30°C; good growth is observed at 22–25°C at pH 7. In TSB agar medium, up to 6% NaCl is tolerated. Cells are positive for catalase, phosphatase, citrate (Simons), lysine and ornithine decarboxylase, and nitrate reduction; but negative for indole production, methyl red, Voges–Proskauer's test, H2S production, gelatinase, lipase, β-galactosidase, and DNase; hydrolyzes urea and starch but not esculin and casein. Produces acid from D-glucose, sucrose, D-fructose, trehalose, rhamnose, D-arabinose, D-galactose, D-xylose, and inositol, but not from erythritol and D-mannose. Utilizes D-glucose, D-galactose, L-rhamnose, L-arabinose, D-raffinose, trehalose, D-melezitose, salicin, sucrose, adonitol, inulin, sodium acetate, glycine, D-alanine, L-glutamic acid, L-proline, L-serine, L-lysine, and L-arginine as sole carbon source but not D-lactose, maltose, D-melibiose, L-sorbose, glycerol, D-cellobiose, mannitol, D-mannose, dulcitol, D-sorbitol, xylitol, erythritol, L-isoleucine, L-threonine, L-aspartic acid, L-glutamate, L-asparagine, methionine, L-tyrosine, L-phenylalanine, tryptophan, and L-histidine. Resistant to (in µg/ml) norfloxacin (25), colistin (25), and nitrofurantoin (30), but sensitive to (in µg/ml) kanamycin (15), ampicillin (25), tetracycline (10), streptomycin (20), and rifampin (15). Diamino acid in the peptidoglycan is lysine, and alanine is present. The major menaquinones present are MK-8 (37%), MK-9 (61%), and MK-10 (2%). Cells contain phosphatidylethanolamine and diphosphatidylglycerol as major polar lipids. The cell-wall sugars are glucose, galactose, and rhamnose. Mycolic acid not present. Major cellular fatty acids are C14:0 (1.30%), C16:0 (2.14%), C18:0 (2.42%), C15:0 iso (11.47%), C15:0 anteiso (54.92%), C15:1 iso (1.25%), C15:1 anteiso (6.38%), C16:0 iso (1.79%), C16:0 anteiso (1.76%), C17:0 anteiso (6.48%), and C19:0 iso (1.07%).
Source: Antarctic sea sediments near the Larsemann Hill area.
DNA G+C content (mol%): 68 ± 0.5 (Tm).
Type strain: SPC26, LMG 24542, NCCB 100228.
Sequence accession no. (16S rRNA gene): AM931709.
Arthrobacter ardleyensis
Chen, Xiao, Wang, Zeng and Wang 2005b, 2235VP (Effective publication: Chen, Xiao, Wang, Zeng and Wang 2005a, 304.)
ard.ley.en'sis. N.L. masc. adj. ardleyensis of or pertaining to Ardley, where the type strain was isolated.
Individual cells show a distinct rod–coccus cycle; some cells are arranged at an angle in V formation, Gram-stain-positive, easily decolorized, rods; motile, aerobic to slightly anaerobic. Catalase is positive, oxidase is negative, and spores or capsules not seen. Colonies in Luria–Bertani medium at 25°C are yellow, circular, convex, smooth, or crumpled in old culture, entire margins, slightly glistening, and semitransparent in young culture. Growth occurs with a suitable carbon source in mineral salts medium; no additional growth factors were required. Grows at 0–30°C; optimal growth temperature is around 25°C. No growth after 30 min at 60°C. Growth is good in 0–3% NaCl and tolerates up to 10% NaCl. Growth is optimal at pH 7.0–8.5. Hydrolyzes Tween 80 and casein but not starch, cellulose, xylan, chitin, and algin. Nitrate is reduced. Methyl red test, Voges–Proskauer test, indole, and H2S production are negative. NH3 production is positive. Arginine dihydrolase is produced, but uricase and tryptophan deaminase are negative. Sensitive to (in µg/ml) ampicillin (100) and chloramphenicol (12.5), but resistant to (in µg/ml) kanamycin (50). A Biolog test showed that the following compounds were utilized for respiration: dextrin, glycogen, Tween 40, Tween 80, L-arabinose, D-arabitol, D-cellobiose, D-fructose, D-galactose, α-D-glucose, D-mannose, maltotriose, maltose, DL-α-glycerolphosphatase, D-ribose, L-rhamnose, D-psicose, palatinose, sucrose, D-tagatose, D-trehalose, turanose, D-xylose, acetic acid, α-hydroxybutyric acid, β-hydroxybutyric acid, γ-hydroxybutyric acid, p-hydroxyphenyl-acetic acid, α-ketoglutaric acid, α-ketovaleric acid, lactamide, D-lactic acid methyl ester, L-lactic acid, D-malic acid, L-malic acid, pyruvic acid methyl ester, succinic acid mono-methyl ester, propionic acid, pyruvic acid, succinamic acid, N-acetyl-L-glutamic acid, L-alaninamide, D-alanine, L-alanine, L-alanyl-glycine, L-asparagine, L-glutamic acid, glycyl-L-glutamic acid, L-pyroglutamic acid, L-serine, putrescine, glycerol, adenosine, and 2′-deoxy adenosine. Negative reactions are observed with α-cyclodextrin, β-cyclodextrin, inulin, mannan, arbutin, amygdalin, N-acetyl-β-D-mannosamine, N-acetyl-D-glucosamine, L-fucose, D-galacturonic acid, gentiobiose, α-D-lactose, lactulose, D-mannitol, D-melezitese, D-melibiose, β-methyl-D-galactoside, 3-methyl-D-glucose, β-methyl-D-glucoside, salicin, stachyose, uridine, uridine-5′-monophosphate, and α-D-glucose 1-phosphate. The interpeptide bridges of the peptidoglycan consist of alanine and glutamic acid. The peptidoglycan type is A4α. The major menaquinones are MK-8 and MK-9. The cellular fatty acid pattern is dominated by C15:0 anteiso.
Source: Antarctic Ardley lake sediment.
DNA G+C content (mol%): 55.2–59.5 (HPLC).
Type strain: An25, CGMCC 1.3685, JCM 12921.
Sequence accession no. (16S rRNA gene; type strain): AJ551163.
Sequence accession no. (16S rRNA gene; other strains): AJ551162, AJ715981.
Arthrobacter arilaitensis
Irlinger, Bimet, Delettre, Lefèvre and Grimont 2005, 459VP
a.ri.lai.ten'sis. N.L. masc. adj. arilaitensis pertaining to Arilait, arbitrary name formed to honor Arilait-Recherches, a research association that coordinates the collective research programmes of the professional French dairy federations.
The description was based on the study of nine strains. Cells are aerobic, Gram-stain-positive, catalase-positive, oxidase-negative, non-spore-forming, nonmotile, and exhibit a rod–coccus growth cycle. Colonies on BHI agar are yellow, round, smooth, convex, and 2 mm in diameter. Grows between 10°C and 30°C and tolerates up to 10% (w/v) NaCl. Gelatinase, β-galactosidase, pyrazinamidase, pyrrolidonyl arylamidase, phosphatase, and α-glucosidase are produced. Urease and esculin are not hydrolyzed. Nitrate is not reduced. Glucose, ribose, xylose, mannitol, maltose, lactose, sucrose, and glycogen are not fermented. In Biotype 100 strips using biotype 2 medium, most strains (>89%) are able to use the following substrates as sole carbon sources: D-glucose, maltotriose, maltose, α-lactose, D-arabitol, glycerol, 5-keto-D-gluconate, D-gluconate, protocatechuate, 4-hydroxybenzoate, lactate, glycerate, and tyrosine. Other substrates are used less frequently (11–88%): D-galactose, sucrose, D-fructose, D-trehalose, D-mannose, lactulose, L-arabinose, D-ribose, D-xylose, malonate, propionate, 2-oxoglutarate, malate, putrescine, succinate, fumarate, D-glucosamine, 3-hydroxybenzoate, 3-hydroxybutyrate, aspartate, glutamate, proline, alanine, L-histidine, serine, methyl-β-galactopyranoside, D-cellobiose, β-gentiobiose, esculin, D-turanose, D-sorbitol, aconitate, citrate, D-glucuronate, 2-keto-D-gluconate, L-tryptophan, phenylacetate, 4-aminobutyrate, caprylate, and 5-aminovalerate. The following carbon sources are not utilized: sorbose, D-melibiose, D-raffinose, methyl-α-galactopyranoside, methyl-β-glucopyranoside, palatinose, L-rhamnose, fucose, D-melezitose, L-arabitol, xylitol, dulcitol, tagatose, myo-inositol, D-mannitol, maltitol, adonitol, lyxose, erythritol, methyl-α-D-glucopyranoside, methyl-D-glucopyranose, saccharate, mucate, tartrate, tricarballylate, D-galacturonate, N-acetyl-D-glucosamine, quinate, gentisate, benzoate, 3-phenylpropionate, m-coumarate, trigonelline, betaine, histamine, caprate, glutarate, ethanolamine, tryptamine, and itaconate. The type strain Re117 utilizes the following substrates as sole carbon sources: D-glucose, D-galactose, D-trehalose, sucrose, maltotriose, maltose, lactose, D-cellobiose, ribose, L-arabinose, D-xylose, D-arabitol, glycerol, turanose, 5-keto-D-gluconate, D-gluconate, protocatechuate, 4-hydroxybenzoate, lactate, glycerate, aspartate, glutamate, alanine, serine, and tyrosine.
The cell wall contains lysine, alanine, and glutamic acid. The whole-cell sugars are galactose, glucose, ribose, and mannose.
Source: the surface of Reblochon cheese.
DNA G+C content (mol%): not determined.
Type strain: Re117, CIP 108037, DSM 16368, JCM 13566.
Sequence accession no. (16S rRNA gene; type strain): AJ609628.
Sequence accession no. (16S rRNA gene; other strains): AJ609621, AJ609622, AJ609623, AJ609624, AJ609625, AJ609626, AJ609627, AJ609628.
Arthrobacter aurescens
(exClark 1951) Phillipps 1953, 241AL (Arthrobacter globiforme var. aurescens Clark 1951, 180)
au.res'cens. L. v. auresco to become golden; L. part. adj. aurescens becoming golden.
Colonies of the type strain are reported to show a yellow pigmentation on agar and other media but this strain gives only pale buff colonies on yeast extract-peptone agar when incubated in the dark. When supplied with biotin, growth occurs in suitable mineral salts medium with an ammonium salt or nitrate as nitrogen source and glucose as carbon and energy source (Keddie et al., 1966; Owens and Keddie, 1969). Starch is hydrolyzed, nitrate is not reduced, and there is no growth in 10% NaCl. Nicotine blue is not produced from nicotine. Utilizes L-arginine, L-asparagine, L-histidine, L-arabinose, D-galactose, D-glucose, D-ribose, D-xylose, butanediol, histidinol, 4-aminobutyrate and ρ-hydroxybenzoate but not L-leucine, L-rhamnose, inositol, or malonate. Assimilates adipic acid, citric acid, formic acid, and uric acid but not benzoic acid, glutaric acid, malonic acid, pimelic acid, or propionic acid. Urea is formed from uric acid. Nonmotile (Kodama et al., 1992). Oxidase-positive. Esculin is hydrolyzed. Elastase but not pyrrolidonyl arylamidase is produced. Gluconate is utilized. The following compounds are utilized for respiration: α-cyclodextrin, D-xylose (weak reaction), arbutin (weak reaction), D-melibiose (weak reaction), 3-methylglucose (weak reaction), and D-raffinose (weak reaction). Uridine, sucrose, propionic acid, and salicin are not used for respiration (Kotoučková et al., 2004). The cell-wall peptidoglycan is of the Lys–Ala–Thr–Ala type (A3α) (Fiedler et al., 1970). The whole-cell sugar is galactose (mannose). The major isoprenoid quinone is MK-9(H2) (Keddie et al., 1986). The predominant fatty acid is C15:0 anteiso (~66%), followed by C17:0 anteiso (~26%). (Kodama et al., 1992)
Source: soil.
DNA G+C content (mol%): 61.5 (Tm).
Type strain: ATCC 13344, CIP 102364, DSM 20116, HAMBI 1850, NBRC 12136, JCM 1330, LMG 3815, NRRL B-2879, VKM Ac-1105.
Sequence accession no. (16S rRNA gene): X83405.
Sequence accession no. (recA): AF214793.
Arthrobacter bergerei
Irlinger, Bimet, Delettre, Lefèvre and Grimont 2005, 459VP
ber.ge're.i. N.L. gen. masc. n. bergerei of Bergère, to honor Jean-Louis Bergère, a French microbiologist.
The description is based on the results of studies of five strains. Cells are aerobic, Gram-stain-positive, catalase-positive, oxidase-negative, non-spore-forming, and nonmotile. They exhibit a rod–coccus growth cycle. Colonies on BHI agar medium are yellow, round, smooth, convex and 2–3 mm in diameter. Grows between 10°C and 30°C and tolerates up to 7.5% (w/v) NaCl. Produce β-galactosidase, pyrazinamidase, pyrrolidonyl arylamidase, and α-glucosidase but not urease, phosphatase, β-glucuronidase, and gelatinase. Esculin is not hydrolyzed. Nitrate is not reduced. D-Glucose, ribose, xylose, mannitol, maltose, lactose, sucrose, and glycogen are not fermented. In Biotype 100 strips using biotype 2 medium, all strains (100%) are able to use the following substrates as sole carbon sources: D-glucose, fructose, D-galactose, sucrose, maltose, lactose, ribose, L-arabinose, D-xylose, D-glycerol, D-gluconate, quinate, protocatechuate, lactate, aspartate, and glutamate. Other substrates are used less frequently (20–80%): lactulose, D-cellobiose, L-rhamnose, D-melezitose, D-mannitol, turanose, D-trehalose, D-mannose, maltotriose, arabitol, methyl-β-galactopyranoside, aconitate, citrate, 2-keto-D-gluconate, L-tryptophan, 4-hydroxybenzoate, 3-hydroxybenzoate, phenylacetate, malate, 5-keto-D-gluconate, betaine, 5-aminovalerate, ethanolamine, malonate, 3-phenylpropionate, coumarate, 4-aminobutyrate, benzoate, putrescine, glucosamine, 3-hydroxybutyrate, histidine, L-alanine, serine, propionate, 2-oxoglutarate, proline, D-alanine, and tyrosine. The following carbon sources are not utilized: sorbose, D-melibiose, D-raffinose, methyl-α-galactopyranoside, β-gentiobiose, methyl-β-glucopyranoside, esculin, palatinose, fucose, L-arabitol, xylitol, dulcitol, tagatose, myo-inositol, maltitol, D-sorbitol, adonitol, lyxose, erythritol, methyl-α-D-glucopyranoside, methyl-D-glucopyranose, saccharate, mucate, D-, L-, and meso-tartrate, tricarballylate, D-glucuronate, D-galacturonate, N-acetyl-D-glucosamine, gentisate, trigonelline, histamine, caprate, caprylate, glutarate, glycerate, tryptamine, and itaconate. The type strain Ca106 utilizes the following substrates as sole carbon sources: D-glucose, fructose, D-galactose, D-mannose, sucrose, lactulose, methyl-β-galactopyranoside, maltotriose, maltose, lactose, D-cellobiose, ribose, L-arabinose, D-xylose, D-arabitol, glycerol, D-gluconate, aconitate, citrate, phenylacetate, quinate, protocatechuate, 3-hydroxybenzoate, benzoate, 4-hydroxybenzoate, putrescine, 4-aminobutyrate, lactate, histidine, glucosamine, aspartate, glutamate, 3-hydroxybutyrate, proline, D-alanine, L-alanine, serine, propionate, and tyrosine. The cell wall contains lysine, alanine, and glutamic acid. The whole-cell sugars are glucose, ribose, and mannose.
Source: the surface of Camembert cheese.
Type strain: Ca106, CIP 108036, DSM 16367, JCM 13567.
Sequence accession no. (16S rRNA gene; type strain): AJ609630.
Sequence accession no. (16S rRNA gene; other strains): AJ609631, AJ609632, AJ609633.
Arthrobacter castelli
Heyrman, Verbeeren, Schumann, Swings and De Vos 2005, 1461VP
cas.tel'li. L. gen. n. castelli of the castle.
Cells are Gram-stain-positive, short rods and cocci (diameter 0.8–1 µm) occurring in pairs and chains. They are nonmotile and do not form endospores. Colonies on NA after 48 h are small (<1 mm), light yellow, round with entire margins, of low convexity, opaque, and smooth. No growth in an anaerobic chamber on NA. Optimal temperature for growth is 22–37°C. Weak growth at 15°C and no growth at 10 or 45°C. Poor growth on media without NaCl. Catalase-positive and oxidase-negative. By using the API CORYNE system, positive reactions are observed for pyrazinamidase, alkaline phosphatase, β-glucuronidase, β-galactosidase, α-glucosidase, and urease. Negative reactions are obtained for nitrate reduction, pyrrolidonyl arylamidase, N-acetyl-β-glucosaminidase, esculin (β-glucosidase), gelatinase, and fermentation of glucose, ribose, xylose, mannitol, maltose, lactose, sucrose, and glycogen. Using the API ZYM system, activity is detected for alkaline phosphatase, acid phosphatase (weak), esterase C4 (weak), esterase lipase C8, lipase C14 (weak), leucine arylamidase, trypsin (weak), phosphoamidase (weak), and α-glucosidase. No activity is detected for valine arylamidase, cystine arylamidase, chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, or α-fucosidase. Predominant fatty acids are C15:0 anteiso (51%) and C15:0 iso (23%). The peptidoglycan type is A3α Lys–Ala–Ser–Ala3. MK-9(H2) is the predominant menaquinone (75%), while MK-10(H2) and MK-9 occur in small amounts. The cell-wall sugars are galactose, xylose, and rhamnose. Polar lipids are phosphatidylinositol, phosphatidylglycerol, diphosphatidylglycerol, one unknown phospholipid, and one unknown glycolipid.
Source: a rosy biofilm overgrowing a mural painting in the Saint-Catherine chapel (castle of Herberstein, Austria).
DNA G+C content (mol%): 68.1 (HPLC).
Type strain: DSM 16402, JCM 21794, LMG 22283.
Sequence accession no. (16S rRNA gene): AJ639826.
Arthrobacter chlorophenolicus
Westerberg, Elväng, Stackebrandt and Jansson 2000, 2090VP
chlo.ro.phe.no'li.cus. N.L. n. chlorophenol chlorophenol; L. masc. suff. -icus suffix used with the sense of pertaining to; N.L. masc. adj. chlorophenolicus pertaining to chlorophenols.
Cells are Gram-stain-positive and display a rod–coccus life cycle. The cocci are ~0.75 µm in diameter. The rods are ~0.5–0.7 µm wide and 1–4 µm, depending on the stage in the life cycle and composition of the growth medium. Agar colonies are circular, smooth, and pearl gray in color. Spores are not formed. Growth occurs with a suitable carbon source in mineral salts medium; no additional growth factors are needed. Red to black pigment is formed from several phenolic compounds, only during incubation with shaking from phenols that can be used for growth, and both with and without shaking for phenols that cannot be used for growth. Catalase-positive. Motile. Obligately aerobic. Grows at 3–37°C and optimally between 20 and 30°C. Grows on acetate, ethanol, glycerol, creatine, uric acid, succinate, glucose, tyrosine, ascorbic acid, nicotine, and weakly on creatinine. Niacin and methanol are not utilized. Testing with Biolog showed that the following compounds are oxidized: dextrin, glycogen, D-fructose, D-galactose, D-gluconic acid, α-D-glucose, maltose, maltotriose, D-mannose, D-melezitose, D-melibiose, D-psicose, D-raffinose, D-ribose, stachyose, sucrose, turanose, acetic acid, β-hydroxybutyric acid, ρ-hydroxyphenylacetic acid, α-ketoglutaric acid, D-lactic acid methyl ester, L-malic acid, methyl pyruvate, pyruvic acid, N-acetyl-L-glutamic acid, D-alanine, L-alanine, L-alanylglycine, L-asparagine, L-glutamic acid, L-pyroglutamic acid, L-serine, and putrescine. The following substrates were not used for respiration: inulin, α-cyclodextrin, β-cyclodextrin, Tween 40,Tween 80, N-acetyl-D-glucosamine, N-acetyl-D-mannosamine, amygdalin, L-arabinose, L-arabitol, arbutin, L-fucose, D-galacturonic acid, α-D-lactose, β-methyl-D-galactoside, α-methyl-D-glucoside, β-methyl-D-glucoside, α-methyl-D-mannoside, salicin, sedoheptulosan, D-tagatose, γ-hydroxybutyric acid, α-ketovaleric acid, lactamide, 2,3-butanediol, 2-deoxyadenosine, inosine, thymidine, adenosine-5′-monophosphate, thymidine-5′-monophosphate, uridine-5′-monophosphate, fructose 6-phosphate, glucose 1-phosphate, glucose 6-phosphate, and D-L-α-glycerol phosphate. The cellular fatty acid content is as follows: C15:0 anteiso (66%), C16:0 iso (10%), C15:0 iso (7%), C17:0 anteiso (5%), C14:0 iso (4%), C16:0 (3%), C14:0 (2%), C16:1 iso h (1%), C16:1 ω7c (1%), and C17:1 anteiso ω9c (1%). The cell-wall diamino acid is lysine. The peptidoglycan type is A3α with an L-Lys–L-Ser–L-Thr–L-Ala interpeptide bridge.
Source: soil at Fort Collins, CO, USA.
DNA G+C content (mol%): 65.1 (Tm).
Type strain: A6, ATCC 700700, CIP 107037, DSM 12829, JCM 12360.
Sequence accession no. (16S rRNA gene): AF102267.
Sequence accession no. (complete genome): CP001341.
Arthrobacter citreus
Sacks 1954, 342AL
ci'tre.us. L. masc. adj. citreus of or pertaining to the citrontree; intended to mean lemon-yellow, lemon-colored.
The rods are feebly motile. Colonies on yeast extract-peptone media have a yellow, nondiffusible pigment which is insoluble in ether and acetone. Biotin, thiamine, nicotinic acid, tyrosine, methionine, cysteine, and a siderophore such as ferrichrome or mycobactin are required for growth. The siderophores can be replaced by certain synthetic metal chelators. The type strain used about 40 of 180 compounds tested as sole or principal sources of carbon + energy. They included a relatively wide range of carbohydrates, sugar derivatives, and amino acids, together with some fatty acids, dicarboxylic acids, hydroxyacids, oxoacids, non-nitrogenous aromatic compounds, amines, and heterocyclic compounds. Of those tested, no simple alcohols, polyalcohols, or glycols were utilized (Keddie et al., 1986). Starch is hydrolyzed, nitrate is reduced, and growth occurs in the presence of 10% NaCl. Nicotine blue is not produced from nicotine. Utilizes L-arginine, L-asparagine, L-histidine, L-arabinose, D-galactose, D-glucose, L-rhamnose, D-ribose, D-xylose, 4-aminobutyrate, and malonate but not L-leucine, butanediol, histidinol, inositol, or ρ-hydroxybenzoate. Assimilate citric acid, formic acid, malonic acid, and propionic acid but not adipic acid, benzoic acid, glutaric acid, pimelic acid, or uric acid. Urea is formed from creatinine but not from uric acid. Motile (only the type strain was investigated; Kodama et al., 1992). The type strain is positive for gelatin-, DNase-, Tween 80-, and tyrosine decomposition, β-galactosidase, and α-glucosidase but not for citrate decomposition, pyrrolidonyl peptidase, or N-acetylglucosaminidase. Acid is produced from glucose and mannitol (Wauters et al., 2000a). The cell-wall peptidoglycan is of the Lys–Thr–Ala2 type (A3α) (Fiedler et al., 1970). The only whole-cell sugar is galactose. The principal isoprenoid quinone is MK-9(H2) (Keddie et al., 1986). The predominant fatty acid is C15:0 anteiso (~57%), followed by C16:0 (~11%), C17:0 anteiso (~8%), C16:0 iso (~7%), and C15:0 iso (~7%) (Kodama et al., 1992).
Source: chicken feces, but probably a dust or soil contaminant.
DNA G+C content (mol%): 62.9–63.8 (Tm).
Type strain: ATCC 11624, CCUG 23840, CIP 102363, DSM 20133, HAMBI 89, NBRC12957, JCM 1331, LMG 16338, NRRL B-1258, VKM Ac-1106.
Sequence accession no. (16S rRNA gene): X80737.
Sequence accession no. (recA): AF214781.
Arthrobacter creatinolyticus
Hou, Kawamura, Sultana, Shu, Hirose, Goto and Ezaki 1998, 428VP
cre.a.ti.no.ly'ti.cus. N.L. n. creatininum creatinine; N.L. masc. adj. lyticus (from Gr. masc. adj. lutikos) able to loosen, able to dissolve; N.L. masc. adj. creatinolyticus creatinine-hydrolyzing.
Grows well aerobically at 37°C, and cells are nonmotile, Gram-stain-positive irregular rods with a length of about 1.4–3.6 µm and a width of 0.5–0.8 µm. Cells change into cocci in older cultures and their diameter is 0.5–0.8 µm. Colonies are circular, smooth, and yellow pigmented on brain heart infusion agar. The diameter of the colonies is about 1–1.5 mm after 24 h culture. The cell-wall amino acid components and ratio are Glu:Ala:Lys, 1.9:3.1:1. The diamino acid is lysine. MK-8 and MK-9 are the major menaquinones. Catalase is produced; creatinine, xanthine, and gelatin but not starch are hydrolyzed; and nitrate is reduced; acid is not produced from xylose, ribose, galactose, rhamnose, arabinose, xylitol, or inositol, but is produced from glycerol.
Source: human urine.
DNA G+C content (mol%): 66.0–67.0 (HPLC).
Type strain: CIP 105749, GIFU 12498, JCM 10102.
Sequence accession no. (16S rRNA gene): D88211.
Arthrobacter crystallopoietes
Ensign and Ritterberg 1963, 149AL
crys.tal.lo.poi.e'tes. L. n. crystallus a crystal, Gr. v. poieo to make, form; N.L. masc. adj. crystallopoietes crystal forming.
Colonies on yeast extract-peptone media show no distinctive pigmentation. On 2-hydroxypyridine agar, a brilliant green crystalline pigment develops in the colony mass after 3 or 4 d incubation at 30°C. Nutritionally nonexacting: growth occurs in a suitable mineral salts medium with an ammonium salt or nitrate as sole nitrogen source and glucose as carbon + energy source. Starch is not hydrolyzed, nicotine is not utilized. Nonmotile (Keddie et al., 1986). Nitrate is reduced. Urease-positive. Esculin is not hydrolyzed. Inositol but not sorbitol, lactose, xylitol, xylose, glucose, maltose, mannitol, and sucrose is utilized (Li et al., 2004c). The cell-wall peptidoglycan is of the Lys–Ala type. The whole-cell sugars are galactose and glucose. The principal isoprenoid quinone is MK-9(H2) (Keddie et al., 1986). The predominant fatty acid is C15:0 anteiso (75%), followed by C17:0 anteiso (7%), C16:0 (4%), C16:0 iso (4%), and C15:0 iso (3%) (Funke et al., 1996).
Source: soil by enrichment in a mineral salt medium containing 2-hydroxypyridine as sole carbon and energy and nitrogen source.
DNA G+C content (mol%): 62.9–63.8 (Bd).
Type strain: ATCC 15481, CIP 102717, DSM 20117, NBRC 14235, JCM 2522, LMG 3819, VKM Ac-1107.
Sequence accession no. (16S rRNA gene): X80738.
Arthrobacter cumminsii
Funke, Hutson, Bernard, Pfyffer, Wauters and Collins 1996, 242VP emend. Funke, Pagano-Niederer, Sjödén and Falsen 1998, 1542 (Effective publication: Funke, Hutson, Bernard, Pfyffer, Wauters and Collins, 1996, 2362.)
cum.min'si.i. N.L. gen. masc. n. cumminsii of Cummins, to honor Cecil S. Cummins, a prominent American microbiologist and a pioneer of chemotaxonomy.
The cells are coryneform bacteria without irregular branching. No spores are formed. The cells are nonmotile. The organism is obligately aerobic. Colonies are whitish-grayish, smooth, slightly convex, and either of creamy or of sticky consistency and are of less than 2 mm in diameter after 24 h of incubation at 37°C in 5% CO2 on SBA. Catalase activity is detected, but nitrate reductase activity is not. Urease activity is variable. Esculinase activity is not detected. DNase and gelatinase activities are observed within 10 d. Acid is produced from ribose but not from maltose, sucrose, mannitol, xylose, lactose, or glycogen. Acid production from glucose is variable. The CAMP reaction is negative. The following other enzyme activities are detected: esterase (C4), esterase lipase (C8), leucine arylamidase, acid phosphatase, and phosphoamidase. The activities of pyrazinamidase, pyrrolidonyl arylamidase, alkaline phosphatase, and cystine arylamidase are variable. Chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase are not present. The peptidoglycan type is L-lysine–L-serine–L-glutamic acid. The predominant fatty acid is C15:0 anteiso. Significant amounts of C15:0 iso, C16:0 iso, C16:0, and C17:0 anteiso are present, depending on the strain. The type strain possesses an A4α, L-lysine–L-serine–L-glutamic acid type murein, but the α-carboxyl group of the D-glutamic acid of the peptide subunit is substituted by a Gly residue.
Source: human clinical specimens.
DNA G+C content (mol%): 60.0–62.0 (HPLC).
Type strain: ATCC 700218, CCUG 36788, CIP 104907, DMMZ 44, DSM 10493, JCM 11675.
Other strains: DMMZ 483, DMMZ 537, CCUG 28802, CCUG 35241, CCUG 36789, CCUG 38876, CCUG 38877, CCUG 38878, CCUG 38880, CCUG 38881, CCUG 38882, CCUG 38883.
Sequence accession no. (16S rRNA gene): X93354.
Arthrobacter defluvii
Kim, Lee, Oh, Kim, Eom and Lee 2008, 1919VP
de.flu'vi.i. L. gen. n. defluvii of sewage.
Cells are aerobic, Gram-stain-positive, nonmotile, non-sporeforming, and display a rod–coccus life cycle. The cocci are approximately 0.6 µm in diameter. The rods are approximately 0.4–0.66 × 1.0–2.0 µm in size. Catalase-positive but oxidase-negative. Growth occurs at 5–37°C (optimum 25–30°C) and at pH 6–10 (optimum pH 7–8). Growth occurs in the presence of up to 5% NaCl but not with 10% NaCl. Colonies are creamy white, translucent, and circular with entire edges. Indole and H2S are not produced. Voges–Proskauer reaction is positive. Nitrate is reduced but nitrite is not. Esculin, casein, and starch (weakly) are hydrolyzed, but gelatin and urea are not. Acid is produced from ribose, D-xylose, inositol, mannitol, and esculin, but not from glycerol, erythritol, D-arabinose, L-arabinose, L-xylose, adonitol, methyl-β-D-xylose, galactose, glucose, fructose, mannose, sorbose, rhamnose, dulcitol, sorbitol, methyl-α-D-mannoside, methyl-α-D-glucoside, N-acetylglucosamine, amygdalin, arbutin, salicin, cellobiose, maltose, lactose, melibiose, sucrose, trehalose, inulin, melezitose, raffinose, starch, glycogen, xylitol, gentiobiose, turanose, D-lyxose, D-tagatose, D-fucose, L-fucose, D-arabitol, L-arabitol, gluconate, 2-ketogluconate, or 5-ketogluconate. The following compounds are utilized as sole carbon sources: mannitol, rhamnose, D-glucose, salicin, D-ribose, melibiose, L-fucose, sucrose, maltose, mannose, L-arabinose, propionate, malonate, valerate, acetate, citrate, gluconate, DL-lactate, malate, phenylacetate, histidine, L-alanine, 2-ketogluconate, 5-ketogluconate, 3-hydroxybutyrate, glycogen, 3-hydroxybenzoate, 4-hydroxybenzoate, L-proline, and L-serine. The following carbon sources are not utilized: N-acetylglucosamine, inositol, D-sorbitol, adipate, itaconate, suberate, and caprate. According to the results from the API ZYM tests, 2-naphthyl butyrate, L-leucyl 2-naphthylamide, 2-naphthyl phosphate (pH 5.4), 6-bromo-2-naphthyl-α-D-galactopyranoside, and 2-naphthyl-α-D-glucopyranoside are hydrolyzed, but 2-naphthyl phosphate (pH 8.5), 2-naphthyl caprylate, 2-naphthyl myristate, L-valyl-2-naphthylamide, L-cystyl-2-naphthylamide, N-benzoyl-DL-arginine-2-naphthylamide, N-glutaryl-phenylalanine 2-naphthylamide, naphthol-AS-BI-phosphate, 2-naphthyl-β-D-galactopyranoside, naphthol-AS-BI-β-D-glucuronide, 6-bromo-2-naphthyl-β-D-glucopyranoside, 1-naphthyl-N-acetyl-β-D-glucosaminide, 6-bromo-2-naphthyl-β-D-mannopyranoside, and 2-naphthyl-α-L-fucopyranoside are not hydrolyzed. The major menaquinone is MK-9(H2); small amounts of MK-8(H2) and MK-7(H2) are also present. Predominant fatty acids are C15:0 anteiso (56.4 ±0.4 %), C16:0 iso (13.7±0.8 %), and C15:0 iso (12.3±2.1 %). Cell-wall peptidoglycan is of A3α type with an L-Lys–L-Ser–L-Thr–L-Ala interpeptide bridge. Cell-wall sugars are galactose, glucose, and rhamnose.
Source: sewage flowing into Geumho River in Daegue, Korea.
DNA G+C content (mol%): 63.5–64.4 (HPLC).
Type strain: 4C1-a, DSM 18782, KCTC 19209.
Sequence accession no. (16S rRNA gene; type strain): AM409361.
Sequence accession no. (16S rRNA gene; another strain): AM409362.
Arthrobacter echigonensis
Ding, Hirose and Yokota 2009, 859VP
echi.go.nen'sis. N.L. masc. adj. echigonensis of or pertaining to the Echigo region, in Japan.
Cells are nonmotile and non-spore-forming. Gram-stain positive, catalase-positive, oxidase-negative. Cells show a rod–coccus growth cycle and produce non-fluorescent pigment. Growth occurs on nutrient broth agar at 10–40°C, optimal temperature for growth is 30°C. Grows in presence of 3–5% NaCl (w/v) but not in the presence of 7% NaCl. The pH range for growth is 6–10 and the optimum pH is 7. Colonies are round, convex, glossy, with entire margins and are light gray or light yellow. Using the API CORYNE system, positive reactions are observed for activities of pyrazinamidase, pyrrolidonyl arylamidase, and for the utilization of glucose, ribose, and sucrose. Negative reactions are obtained for nitrate reduction, hydrolysis of gelatin, urease activity, and for the utilization of xylose, mannitol, lactose, and glycogen. Utilization of maltose is weak. Using the API ZYM system, activity is detected for alkaline phosphatase, acid phosphatase, esterase C4, esterase lipase C8, leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, naphthol-AS-BI-phosphohydrolase β-galactosidase, β-glucuronidase, α-glucosidase, and α-mannosidase. No activity detected for lipase C14, α-chymotrypsin, α-galactosidase, β-glucosidase, N-acetyl-β-glucosaminidase, or α-fucosidase. The predominant fatty acids are C15:0 anteiso, C17:0 anteiso, and C15:0 iso. The diamino acid of the cell-wall peptidoglycan is lysine and the major components are lysine, serine, and alanine. The menaquinone content is MK-9(H2) (83%), MK-10(H2) (6%), and MK-8(H2) (4%).
Source: a filtration substrate made from volcanic rock from Niigata, Japan.
DNA G+C content (mol%): 71.8 (HPLC).
Type strain: LC10, CCTCC AB 206017, IAM 15385, JCM 21829.
Sequence accession no. (16S rRNA gene): AB248531.
Arthrobacter flavus
Reddy, Aggarwal, Matsumoto and Shivaji 2000, 1559VP
fla'vus. L. masc. adj. flavus yellow, the color of a pigment that the bacterium produces.
Cells are aerobic, Gram-stain-positive, non-spore-forming, nonmotile, and non-fermentative; they exhibit a rod–coccus growth cycle. Colonies on peptone-yeast extract medium are yellow, round, smooth, convex, and 0.5–2 mm in diameter. The pigment is insoluble in water but soluble in methanol and has a complex absorption spectrum with maxima at 410, 440, and 470 nm. Pigment production is not dependent on growth conditions or media composition. No growth factors are required. Grows between 5 and 30°C, at pH 6–9, and tolerates up to 11.5% (w/v) NaCl. Optimal growth is observed at 25°C and pH 7. Positive for catalase, lipase, gelatinase, and β-galactosidase but negative for oxidase, urease, phosphatase, indole production, nitrate reduction, and levan formation. Unable to utilize a number of carbon compounds, such as sucrose, rhamnose, cellulose, arabinose, mellibiose, cellobiose, galactose, sucrose, fructose, mannose, trehalose, xylose, mannitol, raffinose, glycerol, ribose, lactose, lactic acid, adonitol, maltose, glucose, glucosamine, sorbitol, melezitol, β-hydroxybutyric acid, dulcitol, glucose, polyethylene glycol, glycine, lysine, sodium citrate, sodium acetate, sodium succinate, cellulose, inulin, meso-inositol, glutamic acid, L-alanine, phenylalanine, methionine, glutamine, arginine, serine, potassium hydrogen phosphate, myristic acid, ammonium formate, creatine, methanol, tyrosine, sodium pyruvate, glycogen, erythritol, tryptophan, ethanol, and sodium thioglycolate. Able to utilize sorbitol as the only source of carbon. Unable to oxidize or ferment glucose, galactose, sucrose, thioglycolate, or mannose but able to acidify sucrose and hydrolyze esculin but not starch or cellulose. Sensitive to all antibiotics (in µg) tested: carbenicillin (50), tobramycin (15), chlortetracycline (30), polymyxin B (300), oxytetracycline (30), rifampin (5), nitrofurantoin (300), penicillin (10), bacitracin (10), nitrofurazone (10), gentamicin (10), lincomycin (2), furazolidone (50), colistin (10), furoxone (100), kanamycin (30), nystatin (100), co-trimoxazole (25), chloramphenicol (30), ampicillin (10), tetracycline (30), amoxicillin (100), trimethoprim (5), and erythromycin (15). The cell-wall peptidoglycan type is Lys–Thr–Ala3 (the A3α variant), and the major menaquinone is MK-9(H2). The cell-wall sugars are galactose, glucose, and ribose.The cellular fatty acids are C14:0, C15:0, C15:0 anteiso, C16:0, C16:0 iso, C16:1, C17:0, C17:0 anteiso, C18:0 iso, C18:0, and C20:0. The polar lipids are phosphatidylglycerol, cardiolipin, and phosphatidylethanolamine.
Source: a cyanobacterial mat sample from McMurdo Dry Valley, Antarctica.
DNA G+C content (mol%): 64.0 ±2.0 (Tm).
Type strain: CMS 19Y, JCM 11496, MTCC 3476.
Arthrobacter gandavensis
Storms, Devriese, Coopman, Schumann, Vyncke and Gillis 2003, 1882VP
gan.da.ven'sis. N.L. masc. adj. gandavensis of or belonging to Gandavum, the Latin name for Ghent, referring to the place where these strains were first isolated.
Cells are Gram-stain-positive, relatively small coccobacilli with one pointed end. They are catalase-positive. Bright yellow pigment is produced. Growth at 25 and 37°C is about equal; growth is less abundant at 42°C. Obligately aerobic. Cells precipitate partially in BHI broth; unable to grow in 6.5% NaCl. Acid is produced weakly from esculin, D-fructose, and ribose. No acid is produced from adonitol, amygdalin, D- or L-arabinose, D- or L-arabitol, arbutin, cellobiose, dulcitol, erythritol, D- or L-fucose, galactose, β-gentiobiose, gluconate, glycogen, glycerol, inositol, inulin, 2- or 5-ketogluconate, lactose, D-lyxose, maltose, maltotriose, mannitol, melezitose, melibiose, methyl-β-glycoside, methyl-α-D-mannoside, methyl-α-D-glucoside, D-raffinose, sucrose, salicin, sorbitol, L-sorbose, starch, D-tagatose, trehalose, D-turanose, xylitol, or D- or L-xylose. Variable in tests with D-glucose, mannose, and rhamnose. Strains are negative for pyrrolidonyl arylamidase [hydrolysis of L-pyroglutamic acid-7-amino-4-methylcoumarin (AMC)], β-glucuronidase, N-acetyl-β-glucosaminidase, enzymic hydrolysis of L-valine-AMC, 4MU-α-D-glucoside, 4-methylumbelliferyl (4MU)-β-D-glucuronide, L-isoleucine-AMC, p-nitrophenyl-β-D-cellobioside, and p-nitrophenyl-α-D-maltoside. Variable in tests for L-arginine-AMC, L-pyroglutamic acid-AMC, L-tryptophan-AMC, 4MU-N-acetyl-β-D-glucosaminide, 4MU-phosphate, 4MU-β-D-glucuronide, and gelatin liquefaction. Cell-wall peptidoglycan is based on the A3α type (L-Lys–L-Thr–L-Ala–L-Ala); major menaquinone is MK-9(H2). Predominant cellular fatty acid is C15:0 anteiso; significant amounts of C15:0 iso and C17:0 anteiso are also present.
Source: mammary and uterine infections in cattle; its pathogenic role in these processes is uncertain.
DNA G+C content (mol%): 65.0 (HPLC).
Type strain: DSM 15046, JCM 13316, LMG 21285.
Other strains: LMG 21286, LMG 21887.
Sequence accession no. (16S rRNA gene; type strain): AJ316140.
Sequence accession no. (16S rRNA; other strains): AJ491107, AJ491108.
Arthrobacter gangotriensis
Gupta, Reddy, Delille and Shivaji 2004, 2376VP
gan.go.tri.en'sis. N.L. masc. adj. gangotriensis of or pertaining to the Indian Antarctic station Dakshin Gangotri.
Cells are aerobic, psychrotolerant, Gram-stain-positive, nonmotile, non-spore-forming, and yellow-pigmented: they exhibit a rod–coccus cycle. Grows between 4 and 30°C. The optimum temperature and pH for growth are 22°C and pH 7. Growth occurs in the presence of 6% NaCl. Positive for catalase, oxidase, phosphatase, urease, and gelatinase and negative for methyl red, indole, and Voges–Proskauer tests, β-galactosidase, arginine dihydrolase, lysine decarboxylase, and arginine decarboxylase. Does not hydrolyze esculin, Tween 80, or starch and does not reduce nitrate to nitrite. Acid is produced from D-fructose, D-galactose, and D-mannose but not from D-arabinose, D-glucose, lactose, D-mannitol, D-rhamnose, D-ribose, sucrose, or D-xylose. Can utilize adonitol, D-arabinose, D-cellobiose, dulcitol, D-galactose, inulin, D-fructose, D-glucose, pyruvate, lactose, D-maltose, D-mannose, D-melibiose, D-ribose, sorbitol, sucrose, D-xylose, xylitol, L-arginine, L-asparagine, L-glycine, and L-phenylalanine but not glycerol, D-mannitol, D-rhamnose, trehalose, L-alanine, L-glutamic acid, L-histidine, L-leucine, or tryptophan as sole carbon sources. Resistant to nalidixic acid and nitrofurantoin but sensitive to amikacin, ampicillin, cefoperazone, cefuroxime, ciprofloxacin, co-trimoxazole, erythromycin, chloramphenicol, kanamycin, lincomycin, lomefloxacin, norfloxacin, penicillin, roxithromycin, streptomycin, tetracycline, tobramycin, and vancomycin. The major menaquinones MK-8, MK-9, and MK-10 are present in the ratio 1:4.5:2. The cellular fatty acids at 25°C are C15:0 anteiso (61.6%), C17:0 anteiso (5.8%), C18:1 (9.0%), C17:0 iso (5.5%), C16:0 iso (4.0%), C15:0 iso (3.0%), and C16:1 (4.2%). The yellow pigment is insoluble in water but soluble in methanol and exhibits three absorption maxima, at 494, 528, and 571.5 nm. The cell-wall peptidoglycan type is Lys–Glu (variation A4α).
Source: penguin rookery soil.
DNA G+C content (mol%): 66.0 (method of analysis specified in Shivaji et al., 1989).
Type strain: DSM 15796; JCM 12166.
Sequence accession no. (16S rRNA gene): AJ606061.
Arthrobacter histidinolovorans
Adams 1954, 832AL
his.ti.di.no.lo.vo'rans. N.L. n. histidinolum histidinol; L. part. adj. vorans devouring, consuming; N.L. part. adj. histidinolovorans histidinol destroying.
Nonmotile. Colonies on yeast extract-peptone media show no distinctive pigmentation. When supplied with biotin, growth occurs in a suitable mineral salts medium with an ammonium salt as sole nitrogen source and glucose as sole carbon and energy source. Utilizes L-histidinol as major source of carbon and energy and nitrogen when grown in mineral salts medium containing a low concentration of yeast extract (Keddie et al., 1986). There is no hydrolysis of starch, no reduction of nitrate, and no growth in 10% NaCl. Nicotine blue is not produced from nicotine. Utilizes L-arginine, L-asparagine, L-histidine, L-arabinose, D-galactose, D-glucose, L-rhamnose, D-ribose, D-xylose, L-histidinol, inositol, 4-aminobutyrate, and ρ-hydroxybenzoate but not L-leucine, butanediol, or malonate. Assimilates citric acid, formic acid, glutaric acid, propionic acid, and uric acid but not adipic acid, benzoic acid, malonic acid, and pimelic acid. Urea is formed from creatinine and uric acid (Kodama et al., 1992). The cell-wall peptidoglycan is of the Lys–Ala–Thr–Ala type (A3α). The whole-cell sugars are galactose and glucose. The principal isoprenoid quinone is MK-9(H2) (Keddie et al., 1986). The predominant fatty acid is C15:0 anteiso (~64%), followed by C17:0 anteiso (~26%) (Kodama et al., 1992).
Source: soil on mineral agar medium containing L-histidinol as carbon and energy and nitrogen source.
DNA G+C content (mol%): 61.3 (Bd).
Type strain: ATCC 11442, CCUG 23888, CIP 106988, DSM 20115, NBRC 15510, JCM 2520, LMG 3822, VKM Ac-1978.
Sequence accession no. (16S rRNA gene): X83406.
Sequence accession no. (recA): AF214788.
Arthrobacter humicola
Kageyama, Morisaki, Ōmura and Takahashi 2008, 56VP
hu.mi'co.la. L. masc. n. humus soil; L. suff. -cola dweller; N.L. masc. or fem. n. humicola soil dweller.
Cells have a rod–coccus cycle. Gram-stain-positive, motile by flagella, and aerobic. Colonies are cream colored on YD agar. Growth occurs on YD agar at initial pH 6–10 and at 4–34°C. In YD agar medium, up to 3% NaCl is tolerated. D-Glucose, D-xylose, raffinose, melibiose, D-mannitol, L-rhamnose, L-inositol, and sucrose are assimilated, but L-arabinose and cellulose are not. Esterase (C4), leucine arylamidase, cystine arylamidase, acid phosphatase, naphthol-AS-BI-phosphohydrolase, α-galactosidase, and α-glucosidase are detected by the API ZYM enzyme assay; the assay is negative for alkaline phosphatase, trypsin, chymotrypsin, β-glucuronidase, N-acetyl-β-glucosaminidase, and α-fucosidase. Weak reactions are detected for esterase lipase (C8), lipase (C14), valine arylamidase, β-galactosidase, β-glucosidase, and α-mannosidase. Catalase is detected by the API Coryne enzyme assay, but nitrate reductase, pyrrolidonyl arylamidase, and urease are negative. The diagnostic diamino acid of the peptidoglycan is lysine. The acyl type of the peptidoglycan is acetyl. The major menaquinone is MK-9(H2). The major cellular fatty acids are C15:0 anteiso, C17:0 anteiso, and C16:0 iso. Cell-wall sugars contain galactose and rhamnose.
Source: paddy soil, Japan.
DNA G+C content (mol%): 67.0 (HPLC).
Type strain: KV-653; JCM 15921, NBRC 102056, NRRL B-24479.
Sequence accession no. (16S rRNA gene): AB279890.
Arthrobacter ilicis
Collins, Jones and Kroppenstedt 1981, 384VP (Effective publication: Collins, Jones and Kroppenstedt 1981, 321.)
i'li.cis. N.L. n. Ilex -icis, a scientific botanical genus name; N.L. gen. n. ilicis, of Ilex.
Surface colonies on nutrient agar are 0.75–1 mm diameter after 1–2 d, becoming larger (2–4 mm) on extended incubation; convex with entire margin, shiny; yellow pigment produced. Cells show a marked change of form during the growth cycle in complete media. Older cultures are composed entirely of largely of coccoid cells which, on transfer to suitable fresh medium, give rise to irregular rods characteristic of exponential phase cultures. Many cells are arranged at an angle to each other to give V-formations. As growth proceeds, the rods become shorter and are eventually replaced by the coccoid cells characteristic of stationary phase cultures (Cure and Keddie, 1973). Does not required B-vitamins: growth occurs in a mineral salts medium containing an ammonium salt and with glucose as carbon and energy source only when Casamino acids are supplied. Gram-stain-positive, non-acid-fast; endospores are not formed. Motile. Optimum growth temperature 25–30°C; growth at 4 and 10°C but not at 37°C. Slight growth in 5% NaCl and no growth in 10% NaCl. Aerobic, catalase- and cytochrome oxidase-positive. No growth anaerobically. Acids are not formed from glucose and other sugars in soli extract media (Jones, 1975). Weak acid production from glycerol after 5 d (Jones, 1975). Methyl red negative. Casein, chitin, gelatin, hippurate, tyrosine, Tween 20, and xanthine hydrolyzed. Urease, phosphatase, and DNase-positive. Esculin, cellulose, starch, nicotine, Tween 60, and Tween 80 are not hydrolyzed. Indole-negative. Sulfatase-negative. Nitrate is not reduced. Sensitive to erythromycin, tetracycline, and streptomycin. Utilizes L-arginine, L-asparagine, L-histidine, L-arabinose, D-galactose, D-glucose, L-rhamnose, D-ribose, D-xylose, histidinol, inositol, 4-aminobutyrate, and ρ-hydroxybenzoate but not L-leucine, butanediol, or malonate. Assimilate citric acid, formic acid, propionic acid, and uric acid but not adipic acid, benzoic acid, glutaric acid, malonic acid, and pimelic acid. Urea is formed from uric acid but not from creatinine (only type strain investigated, Kodama et al., 1992). Elastase and pyrrolidonyl arylamidase are not produced. Does not utilize gluconate. Uses uridine, sucrose, D-melibiose (weak reaction), 3-methylglucose (weak reaction), D-raffinose (weak reaction), and salicin (weak reaction) for respiration but not arbutin and α-cyclodextrin (Kotoučková et al., 2004). The cell-wall peptidoglycan is of the Lys–Ala–Thr–Ala type (A3α). The whole-cell sugars are galactose, rhamnose, and mannose. Mycolic acids are not present. The principal isoprenoid quinone is MK-9(H2). The fatty acid composition is mainly straight-chain, anteiso-, and iso-methyl-branched acids. The major fatty acids are C15:0 anteiso and C17:0 anteiso. The polar lipids comprise diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol, and two glycolipids.
DNA G+C content (mol%): 61.5 (Tm).
Type strain: ATCC 14264, DSM 20138, NCPPB 1228.
Sequence accession no. (16S rRNA gene): X83407.
Additional remarks: Collins et al. (1981) proposed the transfer of Corynebacterium ilicis (Mandel et al., 1961) to the genus Arthrobacter as Arthrobacter ilicis. Several studies however provided evidence that strain ICMP 2608 (= ICPB CI144) and DSM 20138 (= ATCC 14264 = NCPPB 1228) considered to represent the type strain of Corynbacterium ilicis/Arthrobacter ilicis are members of different taxa. Based on evidence from a number of publications, it is clear that DSM 20138 = ATCC 14264 = NCPPB 1228 does not show one of the original diagnostic features of Corynebacterium ilicis, namely pathogenicity on American holly. In contrast, pathogenicity on American holly is exhibited by ICMP 2608 = ICPB CI14 and hence corresponds with the original description of Cornyebacterium ilicis.
After a Request for an Opinion (Young et al., 2004), the Judicial Commission dealt with this problem at its meetings in 2005 at the IUMS Bacteriology and Applied Microbiology Congress in San Francisco. Based on several lines of evidence, the Judicial Commission ruled that the name Arthrobacter ilicis (Collins et al., 1982b) is exemplified by DSM 20138 = ATCC 14264 = NCPPB 1228 and that it is not a plant pathogen according to published evidence. The type strain of Corynebacterium ilicis (Mandel et al., 1961) is represented by the type strain ICMP 2608 = ICPB CI144 and is reported to be a pathogen of American holly. This strain has also been shown to be a member of the species Curtobacterium flaccumfaciens, where it is recognized as the pathotype of Curtobacterium flaccumfaciens pv. ilicis (Tindall, 2008).
Arthrobacter kerguelensis
Gupta, Reddy, Delille and Shivaji 2004, 2376VP
ker.guel.en'sis. N.L. masc. adj. kerguelensis of or pertaining to Kerguelen Islands, Antarctica.
Cells are aerobic, psychrotolerant, Gram-stain-positive, nonmotile, non-spore-forming, and undergo a rod–coccus cycle. Colonies are yellow-pigmented. Grows between 4 and 30°C. The optimum temperature and pH for growth are 22°C and pH 7. Growth occurs in the presence of 6% NaCl. Positive for catalase, oxidase, phosphatase, urease, gelatinase, and lysine decarboxylase but negative for methyl red, indole and Voges–Proskauer tests, β-galactosidase, arginine dihydrolase, and arginine decarboxylase. Hydrolyzes esculin but not Tween 80 or starch, and does not reduce nitrate to nitrite. Acid is produced from D-fructose and D-xylose but not from D-arabinose, D-galactose, D-glucose, lactose, D-mannitol, D-mannose, D-rhamnose, D-ribose, or sucrose. Can utilize adonitol, D-arabinose, dulcitol, D-galactose, inulin, D-fructose, D-glucose, L-glutamic acid, L-histidine, pyruvate, lactose, D-maltose, D-mannose, D-melibiose, D-ribose, sorbitol, sucrose, D-xylose, xylitol, L-arginine, L-asparagine, L-glycine, and L-phenylalanine, D-rhamnose, trehalose but not D-cellobiose, glycerol, D-mannitol, L-alanine, L-leucine, or tryptophan as sole carbon sources. Resistant to nalidixic acid, nitrofurantoin, and norfloxacin but sensitive to amikacin, ampicillin, cefoperazone, cefuroxime, ciprofloxacin, co-trimoxazole, erythromycin, chloramphenicol, kanamycin, lincomycin, lomefloxacin, penicillin, roxithromycin, streptomycin, tetracycline, tobramycin, and vancomycin. The major menaquinones MK-8, MK-9, and MK-10 are present in the ratio 4:6:1. The major cellular fatty acids at 25°C are C15:0 anteiso (50.0%), C17:0 anteiso (25.4%), C15:0 iso (6.7%), C17:0 iso (5.1%), C16:0 iso (4.6%), and C16:1 (3.6%).
Source: sea water.
DNA G+C content (mol%): 58.0 (method of analysis specified in Shivaji et al., 1989).
Type strain: KGN15, DSM 15797, JCM 12165.
Sequence accession no. (16S rRNA gene): AJ606062.
Arthrobacter koreensis
Lee, Lee, Pyun and Bae 2003, 1280VP
ko.re.en'sis. N.L. masc. adj. koreensis of or pertaining to Korea, where the organisms were isolated.
Gram-stain-positive and coryneform. Cells are motile. Colonies on trypticase/soy agar are round, smooth, and yellow. Tests for the reduction of nitrate, the production of catalase, indole, acetoin, pyrazinamidase, alkaline phosphatase, and α-glucosidase, and the assimilation of glucose, mannose, mannitol, maltose, gluconate, and malate are all positive. Oxidizes Tween 40, Tween 80, D-arabitol, D-ribose, xylitol, acetic acid, p-hydroxyphenylacetic acid, methyl pyruvate, pyruvic acid, adenosine, 2′-deoxyadenosine, inosine, thymidine, uridine, and thymidine 5′-monophosphate. Cells are alkalitolerant and do not grow at pH 6.0, but they do grow at pH 7.0–12.0, with an optimum at pH 7.0–8.0. Major isoprenoid quinones are menaquinones MK-8(H2) and MK-9(H2). Major cellular fatty acids are C15:0 anteiso and C15:0 iso. Cell-wall peptidoglycan contains Lys–Thr–Ala2, and the whole-cell sugar is rhamnose.
Source: soil from Daejeon City in Korea.
DNA G+C content (mol%): 63.0 ±2.0 (HPLC).
Type strain: CA15-8, NBRC 16787, JCM 12361, KCTC 9922.
Sequence accession no. (16S rRNA; type strain): AY116496.
Sequence accession no. (16S rRNA; another strain): AY116497.
Arthrobacter luteolus
Wauters, Charlier, Janssens and Delmée 2000b, 1699VP (Effective publication: Wauters, Charlier, Janssens and Delmée 2000a, 2414.)
lu.te'o.lus. L. masc. adj. luteolus yellowish, because of the yellow-pigmented colonies.
Cells of Arthrobacter luteolus are Gram-stain-positive coryneform bacteria. No spores are formed. They are motile by peritrichous flagella. Growth is obligately aerobic. Colonies are slightly yellow, smooth, and ~1.5 mm in diameter after 24 h of incubation at 37°C on blood agar. Catalase is positive. Nitrate is reduced. No urease is detected. Gelatin and tyrosine are hydrolyzed, but not esculin. Simmons' citrate agar is alkalinized. Not susceptible to desferrioxamine. Tween esterase is negative, but DNase is positive. Pyrrolidonyl peptidase is not detected. Acid is produced oxidatively from glucose, maltose, sucrose, and xylose, but not from mannitol and lactose. Glycerol, ribose, D-xylose, D-glucose, D-fructose, D-mannitol, rhamnose, cellobiose, maltose, sucrose, trehalose, xylitol, L-fucose, and 5-keto-gluconate are utilized. Erythritol, D-arabinose, D-arabinose, L-xylose, adonitol, β-methylxyloside, galactose, L-sorbose, dulcitol, inositol, mannitol, sorbitol, α-methyl-D-mannoside, α-methyl-D-glucoside, N-acetylglucosamine, amygdalin, arbutin, esculin, salicin, lactose, melibiose, inulin, melezitose, D-raffinose, starch, glycogen, geniobiose, D-turanose, D-lyxose, D-tagatose, D-fucose, D-arabitol, L-arabitol, and 2-keto-gluconate are not utilized. The following enzymic activities are detected on API ZYM strips: alkaline and acid phosphatase, esterase, esterase-lipase, leucine arylamidase, trypsin, phosphoamidase, and α-glucosidase. Not present are lipase, valine arylamidase, cystine arylamidase, chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, β-glucosidase, N-acetylglucosaminidase, α-mannosidase, and α-fucosidase. The major cellular fatty acid is C15:0 anteiso, and the peptidoglycan type is A3α L-Lys–L-Thr–L-Ala2.
Source: an infected surgical wound.
Type strain: CF25; ATCC BAA-272, CCUG 43811, CIP 106789, DSM 13067, JCM 11676.
Sequence accession no. (16S rRNA gene): AJ243422.
Arthrobacter methylotrophus
Borodina, Kelly, Schumann, Rainey, Ward-Rainey and Wood 2002a, 685VP (Borodina, Kelly, Schumann, Rainey, Ward-Rainey and Wood 2002b, 180)
me.thy.lo.tro'phus. N.L. n. methylum (from French méthyle, back-formation from French méthylène, coined from Gr. n. methu wine and Gr. n. hulê wood) the methyl radical; N.L. pref. methylo-pertaining to the methyl radical; Gr. n. trophos feeder, rearer, one who feeds; N.L. masc. adj. methylotrophus feeding on methyl groups.
Cells are spherical or rod-shaped, showing a rod–coccus growth cycle, 0.6 µm in diameter and 1.3 µm in length. Gram-stain-positive, forming clumps and chains in liquid culture, and are nonmotile; spores or capsules not seen; catalase-and oxidase-positive. Grows heterotrophically and aerobically on glucose, fructose, sucrose, galactose, acetate, ethanol, pyruvate, malate, succinate, citrate, serine, alanine, taurine, and yeast extract, and on alkanesulfonates (propane-, butane-, pentane-, and hexane-sulfonate) and diethylsulfone. Grows aerobically on methylated sulfur compounds (dimethylsulfone, dimethylsulfoxide, dimethylsulfide), methanol, methylamine, trimethylamine, and formaldehyde. Methylotrophic growth with dimethylsulfone uses the ribulose monophosphate cycle for formaldehyde assimilation. Methylotrophic growth with methanol uses the serine pathway for C1-assimilation. Does not grow with methanesulfonate or autotrophically on inorganic sulfur compounds. Nitrate is not reduced. Growth on dimethylsulfone occurs optimally at pH 7.2–7.5 and at 25°C. Temperature range for growth is 4–30°C with no growth at 37 or 44°C. Ammonium chloride, nitrate, methylamine, and EDTA are used as nitrogen sources for growth. Growth occurs in the presence of 1.5% (w/v) NaCl, weakly with 2.5% (w/v) NaCl, but not with 5% (w/v) NaCl. On dimethylsulfone-agar medium, they produce creamy-yellow, circular-convex colonies, 1.0–1.3 mm in diameter. MK-9(H2) is the principal isoprenoid quinone with smaller amounts of MK-10(H2), MK-8(H2), MK-9, MK-7(H2), and MK-11(H2). The principal cellular fatty acid is C15:0 anteiso, with C15:0 iso, C16:0 iso and C17:0 anteiso also present. Peptidoglycan contains lysine as the diagnostic diamino acid, as well as alanine and glutamic acid; provisional peptidoglycan type (L-Lys–L-Ala2–4) is an A11.5, A11.6, or A11.7 structure.
Source: soil from the root system of Tagetes minuta.
DNA G+C content (mol%): 61.0 (Tm).
Type strain: TGA, ATCC BAA-111, DSM 14008, JCM 13519.
Sequence accession no. (16S rRNA gene): AF235090.
Arthrobacter monumenti
Heyrman, Verbeeren, Schumann, Swings and De Vos 2005, 1461VP
mo.nu.men'ti. L. gen. n. monumenti of the monument.
Cells are Gram-stain-positive, short rods and cocci (diameter 0.8–1 µm) occurring in pairs or clusters. They are nonmotile and do not form endospores. Colonies on NA after 48 h are small (<1 mm), light yellow, round with entire margins, of low convexity, opaque and smooth. No growth in an anaerobic chamber on NA. Optimal temperature for growth is 22–30°C. Weak growth at 4°C and 37°C, and no growth at 45°C. Growth on medium with 15% NaCl. Optimum pH for growth is 7–8. Catalase-positive and oxidase-negative. Using the API CORYNE system, positive reactions are observed for nitrate reduction, pyrazinamidase, β-glucuronidase, β-galactosidase, α-glucosidase, esculin (β-glucosidase), and gelatinase. Negative reactions are obtained for N-acetyl-β-glucosaminidase and fermentation of ribose, xylose, mannitol, and glycogen. Variable reactions are obtained for pyrrolidonyl arylamidase, urease, and fermentation of glucose, lactose, maltose, and sucrose. In the variable characters listed above for the API CORYNE tests, the type strain is positive for pyrrolidonyl arylamidase but negative for urease and fermentation of glucose, lactose, maltose, and sucrose. In API ZYM, the type strain is positive for β-galactosidase and α-glucosidase (weak) but negative for α-mannosidase. Using the API ZYM system, activity is detected for alkaline phosphatase, esterase C4, esterase lipase C8, leucine arylamidase, trypsin, acid phosphatase (weak), and phosphoamidase (weak). No activity is detected for lipase C14, valine arylamidase, cystine arylamidase, chymotrypsin, α-galactosidase, β-glucuronidase, β-glucosidase, N-acetyl-β-glucosaminidase, or α-fucosidase. Variable reactions are obtained for β-galactosidase, α-glucosidase, and α-mannosidase. Predominant fatty acids are C15:0 anteiso (57%) and C15:0 iso (25%). The peptidoglycan type is A3α Lys–Ala4. MK-9(H2) is the predominant menaquinone (87%), while MK-7(H2), MK-8(H2), and MK-10(H2) occur in only small amounts. The cell-wall sugars are galactose, xylose, and rhamnose. Polar lipids of the type strain are phosphatidylinositol, phosphatidylglycerol, diphosphatidylglycerol, one unknown phospholipid, and one unknown glycolipid.
Source: a biofilm overgrowing a mural painting in the Servilia tomb (Roman necropolis of Carmona, Spain).
DNA G+C content (mol%): 62.2 (HPLC).
Type strain: DSM 16405, JCM 21770, LMG 19502.
Sequence accession no. (16S rRNA gene): AJ315070.
Arthrobacter mysorens
Nand and Rao 1972, 324AL
my.so'rens. N.L. masc. adj. mysorens of or pertaining to Mysore, India, where the organisms were isolated.
Morphology generally is as for generic description; motility doubtful, flagella not demonstrated. No data available for the peptidoglycan type, cell-wall sugars, or lipids of the type strain.
Growth moderate on nutrient agar; colonies are white and become yellow or lemon-yellow in 3–4 d. Obligately aerobic; weak acid production in glucose, xylose, galactose, glycerol, and cellobiose in 7 d of incubation. Catalase-positive. Growth occurs in Koser's citrate medium; ammonium salts but not nitrate are utilized as sole nitrogen source when provided with glucose as carbon + energy source. No growth on creatine, creatinine, and uric acid agar media; slight growth on nicotine agar. Gelatin and starch hydrolyzed, lecithinase produced. Cellulose not hydrolyzed. Nitrate not reduced to nitrite; urease not produced. Indole, H2S, and acetylmethylcarbinol not produced. Good growth in 10% NaCl broth; optimum temperature between 20–37°C, no growth at 10°C. Glutamic acid is produced in a mineral salts medium when provided with a suitable carbohydrate; a pink water-soluble pigment is produced in the same medium.
Source: sewage samples in Mysore.
DNA G+C content (mol%): not determined.
Type strain: ATCC 33408, CIP 102716, JCM 11565, LMG 16219, NBRC 103060, NCIB (now NCIMB) 10583.
Sequence accession no. (16S rRNA gene): AJ617482.
Additional remarks: Stackebrandt et al. (1983b) carried out detailed study of a strain named Arthrobacter mysorens but unfortunately used ATCC 31021, a patent strain and not the type strain. The authors considered ATCC 31021 to be a member of the Arthrobacter nicotianae group of arthrobacters and to be distinct from those species now recognized.
Arthrobacter nasiphocae
Collins, Hoyles, Foster, Falsen and Weiss 2002a, 571VP
na.si.pho'ca.e. L. masc. n. nasus nose; L. n. phoca seal; N.L. gen. n. nasiphocae of the nose of a seal.
The cells are Gram-stain-positive, non-spore-forming, irregular-shaped rods; coccoid forms may be observed. Colonies are circular, entire, convex, and approximately 1 mm in diameter after 24 h at 37°C on blood agar. Colonies are grayish-white in color and are nonhemolytic on blood agar. Strictly aerobic and catalase-positive. Growth is produced at 25 and 42°C. Grows in broth containing 5% NaCl but not in 10% NaCl. Acid is not produced from glucose, glycogen, lactose, mannitol, maltose, ribose, sucrose, or D-xylose. Activity is detected for alkaline phosphatase, acid phosphatase, α-glucosidase, esterase C4 (weak), phosphoamidase, pyrazinamidase, pyroglutamic acid arylamidase, and leucine arylamidase. No activity is detected for cystine arylamidase, chymotrypsin, ester lipase C8, α-fucosidase, α-galactosidase, β-galactosidase, β-glucosidase, β-glucuronidase, lipase C14, α-mannosidase, N-acetyl-β-glucosaminidase, valine arylamidase, urease, or trypsin. Gelatin and hippurate are hydrolyzed but esculin and starch are not. Nitrate is not reduced. Acetoin is not produced. Cell-wall murein is based on L-lysine variation A3α [type: L-Lys–L-Ala2–Gly2–3–L-Ala (Gly)]. The major menaquinones are MK-9(H2) and MK-8(H2).
Source: the nose of the common seal (Phoca vitulina).
DNA G+C content (mol%): 65.0 (HPLC).
Type strain: M597/99/10, CCUG 42953, CIP 107054, JCM 11677.
Sequence accession no. (16S rRNA gene): AJ292364.
Arthrobacter nicotianae
(Giovannozzi-Sermanni 1959) Stackebrandt, Fowler, Fiedler and Seiler 1983b, 481AL
ni.co.tí.a'na.e. N.L. n. Nicotiana a scientific generic name; N.L. gen. n. nicotianae of Nicotiana, of the tobacco plant.
Growth of the type strain on agar is bright lemon-yellow. The type strain gives abundant growth on nicotine agar: the medium becomes blue at first and deep wine-red later. Nutritionally nonexacting: growth occurs in a suitable mineral salts medium with an ammonium salt as sole nitrogen source (type strain). Nicotine is utilized by the type strain as a sole or major carbon + energy source. More than 70% of the Arthrobacter nicotianae strains assimilate 4-amino-butyrate, 5-amino-valerate, 4-hydroxybenzoate, L-leucine, L-asparagine, D-xylose, d-ribose, L-arabinose, D-galactose, 2,3-butylenglycol, and glycerol; hydrolyze starch and casein; grow in 7% NaCl. The type strain is positive for nitrate reduction, pyrazinamidase, alkaline phosphatase, and pyrrolidonyl arylamidase and assimilates amygdalin, arbutin, cellobiose, D-arbitol, D-xylose, galactose, glycerol, L-arabinose, maltose, ribose, salicin, starch, β-gentiobiose, and glucose. It is negative for β-galactosidase, urease, and cystine arylamidase and cannot assimilate D-mannose, D-turanose, inositol, mannitol, melibiose, rhamnose, sucrose, trehalose, xylitol, N-acetylglucosamine, 5-ketogluconate, and phenylacetate (Margesin et al., 2004; Osorio et al., 1999).
The peptide subunit of the peptidoglycan consists of alanine, D-glutamic acid, and lysine. The interpeptide bridge contains L-alanine and D-glutamic acid (Lys–Ala–Glu type, variation A4α; Fiedler et al., 1970). Cell wall (only the type strain is tested) contains galactose and small amounts of glucose. The long-chain fatty acids are primarily of the iso, anteiso, straight chain, and unsaturated acid types, with the anteiso 12-methyl tetradecanoic acid (C15:0 anteiso) predominating. The major respiratory isoprenoid quinones are unsaturated menaquinones with 8 and 9 isoprene units. Polar lipids consist of diphosphatidylglycerol (DPG), phosphatidylglycerol (PG), and unidentified glycolipids. The type strain contains diglycosyldiacylglycerol with the chromatographic mobility of dimannosyldiacylglycerol (Collins and Kroppenstedt, 1983).
Source: soil and sewage.
DNA G+C content (mol%): 60.0–66.0 (Tm).
Type strain: AS 1.1895, ATCC 15236, CCM 1648, BCRC (formerly CCRC) 11219, CCUG 23842, CDA 883, CIP 82.107, DSM 20123, HAMBI 1859, IAM 12342, NBRC 14234, IMET 10353, JCM 1333, LMG 16305, NCIMB 9458, NRIC 0153.
Sequence accession no. (16S rRNA gene): X80739.
Additional remarks: in DNA–DNA hybridization studies, Stackebrandt et al. (1983b) found reassociation values of 81–93% between the type strain of Arthrobacter nicotianae and the following strains: “Arthrobacter nucleogenes” ATCC 21279, Arthrobacter sp. NCIB 9863, Brevibacterium sp. AJ 1486, and Corynebacterium liquefaciens ATCC 14929. Accordingly they considered that all five strains belong to the same species, Arthrobacter nicotinae.
After continuously decreasing oxygen tension in the growth medium, the type of Arthrobacter nicotianae is able to grow in the presence of nitrate carrying out anaerobic respiratory nitrate reduction (Eschbach et al., 2003).
Arthrobacter nicotinovorans
Kodama, Yamamoto, Amano and Amachi 1992, 237VP
ni.co.ti.no.vo'rans N.L. n. nicotinum nicotine; L. part. adj. vorans devouring, destroying; N.L. part. adj. nicotinovorans nicotine devouring.
Aerobic, Gram-stain-positive, not acid-fast. Cells exhibit a marked rod–coccus growth cycle in complex media. The rods are motile by means of a few lateral flagella. Slight growth occurs in the presence of 10% NaCl. Starch is hydrolyzed, and nitrate is not reduced. Nicotine blue is produced from nicotine. Urea is formed from creatine and uric acid. Utilizes L-arabinose, D-galactose, D-glucose, meso-inositol, D-ribose, D-xylose, 4-aminobutyrate, L-arginine, L-asparagine, L-histidine, and ρ-hydroxybenzoate but not L-leucine. Citric, formic, malonic, uric, and propionic acids are assimilated but glutaric, adipic, pimelic, and benzoic acids are not. The cell-wall peptidoglycan is of the Lys–Ala–Thr–Ala type and the principal isoprenoid quinone is MK-9(H2). The fatty acids are mainly straight chain, anteiso- and isomethyl-branched acids. The major fatty acids are C15:0 and C17:0 anteiso acids.
DNA G+C content (mol%): 62.4 (HPLC).
Type strain: ATCC 49919, CIP 106990, DSM 420, NBRC 15511, JCM 3874, LMG 16253, VKM Ac-1988.
Sequence accession no. (16S rRNA gene): X80743.
Sequence accession no. (recA): AF214788.
Additional remarks: Arthrobacter oxydans DSM 420 was reclassified as Arthrobacter nicotinovorans.
Arthrobacter niigatensis
Ding, Hirose and Yokota 2009, 858VP
ni.i.ga.ten'sis. N.L. masc. adj. niigatensis of or pertaining to the Niigata region, Japan.
Cells are nonmotile and non-spore-forming. Gram-stain positive, catalase-positive, oxidase-negative, shows a rod–coccus growth cycle, and produces non-fluorescent pigment. Growth occurs on nutrient broth agar at 5–40°C, and optimal temperature for growth is 30°C. Grows in the presence of 3–7% (w/v) NaCl. The pH range for growth is 6–11 and the optimum pH is 7.5. Colonies are round, convex, glossy, with entire margins and are light gray or light yellow. Using the API CORYNE system, a positive reaction is observed for reduction of nitrate, activities of pyrazinamidase, pyrrolidonyl arylamidase, and urease, hydrolysis of gelatin, and for the utilization of glucose, ribose, lactose, and sucrose. Maltose is not utilized and mannitol, xylose, and glycogen are weakly utilized. Using the API ZYM system, activity is detected for alkaline phosphatase, acid phosphatase, leucine arylamidase, trypsin, naphthol-AS-BI-phosphohydrolase, esterase C4, esterase lipase C8, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, and α-mannosidase. No activity is detected for lipase C14, α-galactosidase, N-acetyl-β-glucosaminidase, α-chymotrypsin, or α-fucosidase. Activities of valine arylamidase and cystine arylamidase are weak. The predominant fatty acids are C15:0 anteiso, C17:0 anteiso, and C16:0 iso. The diamino acid of the cell-wall peptidoglycan is lysine and major components are lysine, serine, threonine, and alanine. Menaquinones are MK-9(H2) (64%), MK-8(H2) (17%), and MK-8(H4) (15%).
Source: a filtration substrate made from volcanic rock from Niigata, Japan.
DNA G+C content (mol%): 70.8 (HPLC).
Type strain: LC4, CCTCC AB 206012, IAM 15382, JCM 21826.
Sequence accession no. (16S rRNA gene): AB248526.
Arthrobacter nitroguajacolicus
Kotoučková, Schumann, Durnová, Spröer, Sedláček, Neča, Zdráhal and Němec 2004, 776VP
ni.tro.gu.a.ja.co'li.cus. N.L. masc. adj. nitroguajacolicus pertaining to the chemical nitroguaiacol, whose name is based on N.L. guaiacum from Spanish guayaco, a tropical American tree and the resin derived thereof.
Cells are Gram-stain-positive irregular rods, club-shaped with typical V-forms, motile, and non-acid-fast. They display a rod–coccus life cycle. Cocci are 0.7–1 µm in diameter; rods are 0.6–1.0 µm wide and 1.0–4 µm long. Spores are not formed. Colonies are yellow, circular, convex, and opaque. Growth occurs with a suitable carbon source in mineral salts medium; no additional growth factors are required. Obligately aerobic. Growth at 4–37°C, with optimum at 25–30°C. Growth occurs in the pH range 6.0–8.0 and in the presence of up to 6% (w/v) NaCl. Catalase- and oxidase-positive. Nitrate not reduced. Methyl red test, urease, and hemolysis negative. Hydrolyzes gelatin, starch, casein, esculin, o-nitrophenyl-β-D-galactopyranoside, and tyrosine are hydrolyzed. Tween 80, DNA, and lecithin are not hydrolyzed. Elastase is produced. Pyrrolidonyl arylamidase and arginine dihydrolase are not produced. Production of alkaline phosphatase, acid phosphatase, α-galactosidase, and α-fucosidase is variable between strains: the type strain shows positive reactions. Simmons' citrate is utilized, but not gluconate. Acid is not produced from ribose, mannitol, sorbitol, lactose, trehalose, raffinose, sucrose, L-arabinose, D-arabitol, cyclodextrin, glycogen, pullulan, maltose, melibiose, melezitose, methyl-β-D-glucopyranoside, or tagatose. In the Biolog test system, the following compounds are utilized for respiration: dextrin, glycogen, arbutin, D-cellobiose, D-fructose, D-galactose, α-D-glucose, maltose, maltotriose, D-mannitol, D-mannose, palatinose, D-psicose, D-raffinose, D-ribose, D-sorbitol, sucrose, turanose, acetic acid, α-hydroxybutyric acid, β-hydroxybutyric acid, p-hydroxyphenylacetic acid, L-malic acid, methyl pyruvate, propionic acid, pyruvic acid, alaninamide, D-alanine, L-alanine, L-alanyl-glycine, L-asparagine, L-glutamic acid, glycyl-L-glutamic acid, L-pyroglutamic acid, L-serine, putrescine, and glycerol. Utilization of gentiobiose, α-ketoglutaric acid and β-ketoglutaric acid is strain dependent: the type strain is positive. A negative reaction is seen with α-cyclodextrin, β-cyclodextrin, inulin, mannan, Tween 40, Tween 80, N-acetyl-D-glucosamine, N-acetyl-D-mannosamine, amygdalin, L-arabinose, D-arabitol, L-fucose, D-galacturonic acid, α-D-lactose, lactulose, D-melezitose, D-melibiose, methyl-α-D-galactoside, methyl-β-D-galactoside, 3-methylglucose, methyl-α-D-glucoside, methyl-β-D-glucoside, methyl-α-D-mannoside, L-rhamnose, salicin, sedoheptulosan, D-tagatose, D-trehalose, xylitol, D-xylose, γ-hydroxybutyric acid, lactamide, D-lactic acid methyl ester, D-malic acid, monomethyl succinate, succinamic acid, succinic acid, N-acetyl glutamic acid, 2,3-butanediol, adenosine, 2-deoxyadenosine, inosine, thymidine, uridine, adenosine 5′-monophosphate, thymidine 5′-monophosphate, uridine 5′-monophosphate, fructose 6-phosphate, glucose 1-phosphate, glucose 6-phosphate, and DL-α-glycerol phosphate. The cell-wall diamino acid is lysine. The peptidoglycan type is A3α, with an Ala–Thr–Ala interpeptide bridge. The major menaquinone is MK-9(H2); MK-8(H2) and MK-10(H2) occur as minor components. The cellular fatty acid pattern is dominated by C15:0 anteiso (65–70%).
Source: forest soil.
DNA G+C content (mol%): 61.9 (HPLC).
Type strain: G2-1, CCM 4924, DSM 15232, JCM 14115.
Other strains: CCM 4925, CCM 7049.
Sequence accession no. (16S rRNA gene): AJ512504.
Arthrobacter oryzae
Kageyama, Morisaki, Ōmura and Takahashi 2008, 55VP
o.ry'za.e. L. gen. n. oryzae of rice.
Cells have a rod–coccus cycle. Gram-stain-positive, motile by flagella and aerobic. Colonies are cream colored on YD agar. Growth occurs on YD agar at initial pH values between 6 and 11 and at temperatures between 4 and 34°C. In YD agar medium, up to 2% NaCl is tolerated. D-Glucose, raffinose, melibiose, D-mannitol, L-inositol, and sucrose are assimilated, but L-arabinose and cellulose are not. Leucine arylamidase, acid phosphatase, naphthol-AS-BI-phosphohydrolase, β-glucuronidase, and α-glucosidase are detected by the API ZYM enzyme assay; the assay is negative for alkaline phosphatase, esterase lipase (C8), trypsin, chymotrypsin, α-galactosidase, β-galactosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase. Weak reactions are detected for esterase (C4), lipase (C14), valine arylamidase, and cystine arylamidase. Nitrate reductase, pyrrolidonyl arylamidase, and catalase are detected by the API Coryne enzyme assay, but urease is negative. The diagnostic diamino acid of the peptidoglycan is lysine. The acyl type of the peptidoglycan is acetyl. The major menaquinone is MK-9(H2). The major cellular fatty acids are C15:0 anteiso, C17:0 anteiso, and C15:0 iso. Cell-wall sugars contain galactose and glucose.
Source: paddy soil, Japan.
DNA G+C content (mol%): 67.0 (HPLC).
Type strain: KV-651, JCM 15922, NBRC 102055, NRRL B-24478.
Sequence accession no. (16S rRNA gene): AB279889.
Arthrobacter oxydans
Sguros 1954, 21AL
o.xy'dans. N.L. part. adj. oxydans oxidizing.
Nonmotile (only type strain investigated; Kodama et al., 1992; Li et al., 2004c). Colonies on yeast extract-peptone media are either pearl-gray/white or show a yellow, nondiffusible pigment depending on strain. Growth on nicotinemineral salts-yeast extract agar is abundant with production of a deep blue, diffusible pigment which turns reddish or yellow-brown in older cultures. When supplied with biotin, growth occurs in a suitable mineral salts medium with an ammonium salt as nitrogen source and glucose as carbon and energy source (Keddie et al., 1986). Starch is hydrolyzed, nitrate is reduced, grows in 10% NaCl. Nicotine blue is produced from nicotine. Esculin is not hydrolyzed. Utilizes L-arginine, L-asparagine, L-histidine, L-leucine, L-arabinose, D-galactose, D-glucose, L-rhamnose, D-ribose, D-xylose, butanediol, histidinol, inositol, malonate, sorbitol, lactose, xylitol, and sucrose but not maltose. Assimilates citric acid, formic acid, propionic acid, and uric acid but not adipic acid, benzoic acid, glutaric acid, malonic acid, or pimelic acid. Urea is formed from creatinine and uric acid. Gelatin and tyrosine are hydrolyzed. Positive for DNase, citrate, β-galactosidase, and α-glucosidase and negative for pyrrolidonyl peptidase and N-acetylglucosaminidase. Acid is formed from glucose (Wauters et al., 2000a). Wauters et al. (2000a) described the formation of acid from mannitol, while Li et al. (2004c) showed that the type strain of Arthrobacter oxydans does not utilize mannitol. The cell-wall peptidoglycan is of the Lys–Ser–Thr–Ala type (A3α). The whole-cell sugars are galactose and glucose. The principal isoprenoid quinone is MK-9(H2) (Keddie et al., 1986). The predominant fatty acid is C15:0 anteiso (~45%), followed by C17:0 anteiso (~17%), C16:0 iso (~11%), C16:0 (~11%), and C15:0 iso (~9%) (only type strain investigated; Kodama et al., 1992).
Source: cured tobacco leaves and associated air. Isolated by enrichment in nicotine-mineral salts-yeast extract medium.
DNA G+C content (mol%): 62.7–64.4 (Tm).
Type strain: AS 1.1925, ATCC 14358, BCRC (formerly CCRC) 11573, CCUG 17757, CIP 107005, DSM 20119, HAMBI 1857, NBRC 12138, IMET 10684, JCM 2521, LMG 3816, NCIMB 9333, NRIC 0154, VKM Ac-1114.
Sequence accession no. (16S rRNA gene): X83408.
Sequence accession no. (recA): AF214789.
Arthrobacter parietis
Heyrman, Verbeeren, Schumann, Swings and De Vos 2005, 1462VP
pa.ri'e.tis. L. gen. n. parietis of a wall.
Cells are Gram-stain-positive, short rods and cocci (diameter 0.8–1 µm) occurring in pairs or clusters. They are non-motile and do not form endospores. Colonies on NA after 48 h are 1–2 mm in diameter, yellow-orange, round with entire margins, of low convexity, opaque, and smooth. No growth in an anaerobic chamber on NA. Optimal temperature for growth is 22–30°C. No or only weak growth at 37°C and no growth at 45°C. Good growth after 1 week at 4°C. Growth on medium with 15% NaCl. Growth occurs at pH 6–9, with an optimum of 7–8. Catalase-positive and oxidase-negative. Using the API CORYNE system, positive reactions are observed for nitrate reduction, pyrazinamidase, β-galactosidase, α-glucosidase, esculin (β-glucosidase), gelatinase, and fermentation of glucose. Negative reactions are obtained for alkaline phosphatase, β-glucuronidase, N-acetyl-β-glucosaminidase and fermentation of ribose, xylose, mannitol, and glycogen. Variable reactions are obtained for pyrrolidonyl arylamidase, urease, and fermentation of maltose, lactose, and sucrose. Using the API ZYM system, activity is detected for acid phosphatase (weak), esterase C4, leucine arylamidase, trypsin, phosphoamidase (weak), α-galactosidase, β-galactosidase, and α-glucosidase. No activity is detected for alkaline phosphatase, esterase lipase C8, lipase C14, valine arylamidase, chymotrypsin, β-glucuronidase, N-acetyl-β-glucosaminidase, and α-fucosidase. Variable results (if positive, weak) were obtained for cystine arylamidase, β-glucosidase, and α-mannosidase. The type strain is negative for the variable characters listed above. Predominant fatty acids are C15:0 anteiso (51%) and C15:0 iso (29%). The peptidoglycan type is A3α Lys–Thr–Ala2. MK-9(H2) (69%) and MK-10(H2) (20%) are the predominant menaquinones, while MK-8(H2) and MK-11(H2) occur in only small amounts. The cell-wall sugar is galactose. Polar lipids of the type strain are phosphatidylinositol, phosphatidylglycerol, diphosphatidylglycerol, one unknown phospholipid, and two unknown glycolipids.
Source: a biofilm overgrowing a mural painting in the Servilia tomb (Roman necropolis of Carmona, Spain).
DNA G+C content (mol%): 63.8 (HPLC).
Type strain: DSM 16404, JCM 14917, LMG 22281.
Sequence accession no. (16S rRNA gene): AJ639830.
Arthrobacter pascens
Lochhead and Burton 1953, 7AL
pas'cens. L. part adj. pascens nourishing.
Colonies on yeast-peptone medium show no distinctive pigmentation. Nutritionally nonexacting: growth occurs in a suitable mineral salts medium with an ammonium salt or nitrate as sole nitrogen source and glucose as carbon + energy source. Starch is hydrolyzed, nicotine is not utilized. Nonmotile. The cell-wall peptidoglycan is of the Lys–Ala2 type (A3α) (Fiedler et al., 1970). The whole-cell sugars are galactose and glucose. The principal isoprenoid quinone is MK-9(H2). The predominant fatty acid is C15:0 anteiso (59%), followed by C17:0 anteiso (13%), C15:0 iso (6%), C16:0 iso (3%), C16:0 (3%), C16:1 ω7c (3%), and C18:1 ω9c (2%) (Funke et al., 1996).
Source: soil.
DNA G+C content (mol%): 63.7 (Tm).
Type strain: ATCC 13346, CCUG 23843, CIP 102362, DSM 20545, HAMBI 1862, NBRC 12139, JCM 11606, LMG 16255, NRRL B-1814, VKM Ac-1116.
Sequence accession no. (16S rRNA gene): X80740.
Sequence accession no. (recA): AF214789.
Arthrobacter phenanthrenivorans
Kallimanis, Kavakiotis, Perisynakis, Spröer, Pukall, Drainas and Koukkou 2009, 278VP
phe.nan.thre.ni.vo'rans. N.L. n. phenanthrenum phenanthrene; L. v. vorare to devour; L. part. adj. vorans devouring, digesting; N.L. part. adj. phenanthrenivorans digesting phenanthrene.
Cells are aerobic and nonmotile, stain Gram positive, and exhibit a rod–coccus growth cycle. Colonies are cream to yellow in color. Grows at 4–37°C in mineral salts medium with a suitable carbon source; optimum growth occurs between 30 and 37°C. No additional growth factors are required. Catalase- and amylase-positive. Reduces nitrate to nitrite. Negative for oxidase, urease, lipase, and gelatinase. Does not produce H2S. Acid is not produced from glucose, lactose, or sucrose. Utilizes phenanthrene and anthracene as sole carbon sources. The polar lipid pattern consists of phosphatidylethanolamine, phosphatidylglycerol, and diphosphatidylglycerol. The major menaquinone is MK-8 and the major fatty acids are C15:0 anteiso, C16:0 iso, C15:0 iso, C16:0, and C17:0 anteiso.
Source: creosote-contaminated soil in Greece.
DNA G+C content (mol%): 65.7 ±0.2 (Tm).
Type strain: Sphe3, DSM 18606, JCM 16027, LMG 23796.
Sequence accession no. (16S rRNA gene): AM176541.
Sequence accession no. (recA): AM931439.
Arthrobacter pigmenti
Heyrman, Verbeeren, Schumann, Swings and De Vos 2005, 1462VP
pig.men'ti. L. gen. n. pigmenti of pigment or paint.
Cells are Gram-stain-positive, short rods and cocci (diameter 0.8–1 µm) occurring in pairs, chains, or clusters. They are nonmotile and do not form endospores. Colonies on NA after 48 h are small (<1 mm), light yellow, round with entire margins, of low convexity, opaque, and smooth. No growth in an anaerobic chamber on NA. Optimum temperature for growth is 22–30°C. No or only weak growth at 37 or 10°C. No growth at 45 or 4°C. Growth on medium with 15% NaCl; optimal growth at 10% NaCl. Grows at pH 7–10 and optimally at pH 8–9. Catalase-positive and oxidase-negative. Using the API CORYNE system, positive reactions are observed for pyrazinamidase, β-glucuronidase, β-galactosidase, α-glucosidase, and gelatinase. Negative reactions are obtained for pyrrolidonyl arylamidase, N-acetyl-β-glucosaminidase, urease, and fermentation of ribose, xylose, mannitol, lactose, and glycogen. Variable reactions are obtained for nitrate reduction, esculin (β-glucosidase) and fermentation of glucose, maltose, and sucrose. Using the API ZYM system, activity is detected for alkaline phosphatase, esterase lipase C8 (weak), leucine arylamidase, trypsin, phosphoamidase (weak), α-galactosidase (weak), and β-glucuronidase. No activity is detected for lipase C14, valine arylamidase, cystine arylamidase, chymotrypsin, α-galactosidase, β-glucosidase, N-acetyl-β-glucosaminidase, or α-fucosidase. Variable results (if positive, weak) were obtained for esterase C4, acid phosphatase, and α-mannosidase. Predominant fatty acids are C15:0 iso (48%) and C15:0 anteiso (40%). The peptidoglycan type is A3α Lys–Ala4. MK-9(H2) (66%), MK-10(H2) (13%), and MK-7(H2) (14%) are the predominant menaquinones, while MK-9 occurs in only small amounts. The cell-wall sugars are galactose and rhamnose. Polar lipids of the type strain are phosphatidylinositol, phosphatidylglycerol, diphosphatidylglycerol, one unknown phospholipid, and one unknown glycolipid.
Source: a rosy biofilm overgrowing a mural painting in the Saint-Catherine chapel (castle of Herberstein, Austria).
DNA G+C content (mol%): 61.6 (HPLC).
Type strain: DSM 16403, JCM 21771, LMG 22284.
Sequence accession no. (16S rRNA gene): AJ639827.
Arthrobacter polychromogenes
Schippers-Lammertse, Muijsers and Klatser-Oedekerk 1963, 2AL
po.ly.chro.mo'ge.nes. Gr. adj. polu many; Gr. n. chroma color; Gr. v. gennaio produce; N.L. part. adj. polychromogenes producing many colors.
Cells are nonmotile, long rods and cocci. On nutrient agar, colonies are white, circular, smooth, convex, with an entire edge. Colonies are blue colored on peptone-glucose agar due to large amounts of blue-black crystals in the colonies. Gelatin and starch are hydrolyzed. Negative for indole production, hydrogen sulfide production, acetoin production, and urease. Little or no acid from sugars. Acid from glycerol. Nitrate is reduced to nitrite. Nicotine blue is not produced from nicotine. Utilizes L-arginine, L-asparagine, L-histidine, L-leucine, L-arabinose, D-galactose, D-glucose, L-rhamnose, D-ribose, D-xylose, butanediol, and malonate but not inositol. Assimilates citric acid, formic acid, propionic acid, and uric acid but not adipic acid, benzoic acid, glutaric acid, malonic acid, or pimelic acid (Kodama et al., 1992).
Ammonium or nitrate without biotin is not sufficient as nitrogen source. Sodium citrate is sufficient as sole carbon source, also in the absence of biotin. Excellent growth and much pigment is obtained on a medium of peptone 1%, glycerol 2%, KCl 0.6%, agar 2%, pH 9.0. The bacterium also grows well, though is colorless, when vitamin-free Casamino acids are substituted for peptone in the last mentioned medium; the bacterium does not need vitamins for growth. The color appears in the vitamin-free medium after addition of biotin; biotin is necessary for the formation of the pigments. The temperature optimum for growth and pigment production is about 25°C. Most often growth but no color is observed at 37°C. Growth at 10°C but not at 41°C. A colorless culture, grown at 37°C, soon becomes colored at room temperature. At a pH of about 5, no growth occurs. Optimum pigment formation occurs at the rather high pH value of 9–10. The maximum pH for growth and pigment production is 11 or above. No pigment formation at a pH below 6. The cell-wall peptidoglycan is of the Lys–Ser–Thr–Ala type (A3α) (Fiedler et al., 1970). The whole-cell sugar is galactose. The principal isoprenoid quinone is MK-9(H2). The predominant fatty acid is C15:0 anteiso (~44%), followed by C17:0 anteiso (~25%), C16:0 iso (~6%), C16:0 (~13%), C16:0 iso (~6%), and C15:0 iso (~3%) (Huang et al., 2005b; Kodama et al., 1992).
Source: air.
DNA G+C content (mol%): 62.9 (Tm; Kodama et al., 1992).
Type strain: ATCC 15216, CIP 106989, DSM 20136, NBRC 15512, JCM 2523, LMG 3821, VKM Ac-1955.
Sequence accession no. (16S rRNA gene): X80741.
Arthrobacter protophormiae
(ex Lysenko 1959) Stackebrandt, Fowler, Fiedler and Seiler 1984, 270VP (Effective publication: Stackebrandt, Fowler, Fiedler and Seiler 1983b, 482.)
pro.to.phor.mi'a.e. N.L. n. Protophormia a genus of dipteran insects, N.L. gen. n. protophormiae of Protophormia.
In 1-d-old cultures, the cells are slightly club-shaped rods with rounded ends (0.6–0.8 × 0.8–2.0 µm) occurring singly or in irregular groups. Stationary-phase cultures (older than 2 d) are composed almost entirely of spherical cells only. Colonies on nutrient agar show a pale to sulfur yellow, nondiffusible pigment. The peptide subunit of the peptidoglycan consists of alanine, D-glutamic acid, and lysine. The interpeptide bridge contains alanine and glutamic acid (Lys–Ala–Glu type, variation A4α).
Cell wall (only the type strain is tested) contains galactose and small amounts of glucose. The long-chain fatty acids are primarily of the iso, anteiso, straight chain, and unsaturated types, with the anteiso 12-methyl tetradecanoic acid (C15:0 anteiso) predominating. The major respiratory isoprenoid quinones are unsaturated menaquinones with 8 and 9 isoprene units (MK-8 and MK-9). Polar lipids consist of diphosphatidylglycerol (DPG), phosphatidylglycerol (PG), and unidentified glycolipids. More than 80% of the 6 strains of Arthrobacter protophormiae strains assimilate 5-amino-valerate, 4-hydroxybenzoate, L-asparagine, L-arginine, L-histidine, L-arabinose, D-galactose, and glycerol; resistant to 7% NaCl.
Source: the fly Protophormia terraenovae and the soil.
DNA G+C content (mol%): 63.2–65.9 (Tm).
Type strain: ATCC 19271, CIP 106987, DSM 20168, NBRC 12128, JCM 1973, LMG 16324, VKM Ac-2104.
Sequence accession no. (16S rRNA gene): X80745.
Sequence accession no. (recA): AF214790.
Arthrobacter psychrochitiniphilus
Wang, Gai, Chen and Xiao 2009, 2761VP
psy.chro.chi.ti.ni'phi.lus. Gr. adj. psuchros cold; N.L. n. chitinum chitin; Gr. adj. philos loving; N.L. masc. adj. psychrochitiniphilus a cold, chitin-loving bacterium.
Individual cells show a distinct rod–coccus cycle. Cells are Gram-stain-positive, aerobic, catalase-positive, and oxidase-negative motile rods. Spores or capsules are not seen. Colonies in LB medium at 20°C are yellow, circular, and convex. Growth occurs at 0–25°C, the optimal growth temperature is around 20°C. Grows well at 0–3% NaCl. Optimal growth occurs at pH 6–8. Tween 80, starch, cellulose, lactose, and chitin are hydrolyzed. Gelatin, lecithin, and urea are not hydrolyzed. Nitrate is reduced and NH3 production is positive. Production of indole and H2S is negative. Sensitive to ampicillin, chloramphenicol, kanamycin, streptomycin, and tetracycline. Growth occurs on lactose or chitin as the sole carbon source. The cellular fatty acid pattern is dominated by C15:0 anteiso. The peptidoglycan type is A3α. The major menaquinone is MK-9(H2). Biolog tests show that the following compounds are utilized for respiration: dextrin, Tween 40, Tween 80, N-acetyl-D-glucosamine, L-arabinose, D-arabitol, cellobiose, D-fructose, D-galactose, D-glucose, α-D-lactose, lactulose, maltose, maltotriose, D-mannitol, D-mannose, D-melezitose, melibiose, methyl-β-D-galactoside, D-psicose, D-ribose, D-sorbitol, sucrose, D-tagatose, trehalose, turanose, xylitol, D-xylose, acetic acid, α-hydroxybutyric acid, α-ketovaleric acid, D-lactic acid methyl ester, L-lactic acid, L-malic acid, pyruvic acid methyl ester, succinic acid monomethyl ester, propionic acid, pyruvic acid, succinamic acid, succinic acid, N-acetyl-L-glutamic acid, D-alanine, L-alanyl glycine, L-asparagine, L-glutamic acid, glycyl L-glutamic acid, L-serine, putrescine, 2,3-butanediol, glycerol, adenosine, inosine, thymidine, uridine, adenosine 5′-monophosphate, thymidine 5′-monophosphate, uridine 5′-monophosphate, D-glucose 6-phosphate, and DL-α-glycerol phosphate.
Source: guano of Adelie penguins, Antarctica.
DNA G+C content (mol%): 58.5 (HPLC).
Type strain: GP3, CGMCC 1.6355, JCM 13874.
Arthrobacter psychrolactophilus
Loveland-Curtze, Sheridan, Gutshall and Brechley 2000, 3VP (Effective publication: Love-land-Curtze, Sheridan, Gutshall and Brechley 1999, 362.)
psy.chro.lac.to'phi.lus. Gr. adj. psuchros cold; L. n. lac lactis milk; Gr. masc. adj. philos friend, loving; N.L. masc. adj. psychrolactophilus a cold, milk (sugar)-loving (bacterium).
Individual cells show a distinct rod–coccus cycle and have a mean cell length of 1.4 µm during exponential growth and 0.5 µm during stationary phase. Gram-stain-positive; easily decolorized; strict aerobe. Contains lysine as the diagnostic amino acid in the peptidoglycan. Non-spore-forming. Nonmotile. No vitamin requirements; grows in mineral salts medium with ammonium chloride as sole source of nitrogen. Colonies on trypticase soy agar without glucose are yellow; degree of pigmentation varies with growth temperature and age of the cells. Able to grow at 0–5°C. Growth range, 0–30°C. Generation time at 10°C in trypticase soy broth with no added carbohydrate is 4.8 h. Does not produce acid with glucose as carbon source. Can utilize lactose, sorbitol, melibiose, cellobiose, glycerol, maltose, raffinose, xylose, galactose, sucrose, and glucose as sole carbon sources. Produces catalase. Produces β-galactosidase, β-glucuronidase, β-glucosidase, β-glucosidase, amylase, and gelatinase. Negative for nitrate reduction, alkaline phosphatase, N-acetylglucosaminidase, urease, and DNase. The major cellular fatty acids are anteiso- and isobranched fatty acids. The predominant fatty acid is C15:0 anteiso (~73%), followed by C17:0 anteiso (~13%), C16:0 iso (~8%), C16:0 (~2%), and C15:0 iso (~1.4%). All other fatty acids are at levels below 1%.
Source: soil.
DNA G+C content (mol%): 60.6 (Tm).
Type strain: B7, ATCC 700733, JCM 12399.
Sequence accession no. (16S rRNA gene): AF134179.
Arthrobacter psychrophenolicus
Margesin, Schumann, Spröer and Gounot 2004, 2070VP
psy.chro.phe.no'li.cus. Gr. adj. psuchros cold; N.L. masc. adj. phenolicus relating to phenol; N.L. masc. adj. psychrophenolicus relating to phenol [degradation] at low temperatures.
On nutrient agar, colonies are round, convex, glossy, and have entire margins and a yellow, non-fluorescent pigment. Cells are Gram-stain-positive, aerobic, non-spore-forming, and nonmotile. They exhibit a rod–coccus cycle with irregular rods in the exponential growth phase and predominantly coccoid cells in the stationary growth phase. Positive for phenol degradation, nitrate reduction, and urease, but negative for alkaline phosphatase, pyrrolidonyl arylamidase, and cystine arylamidase. Hydrolyze skimmed milk at 10°C and 25°C. Can utilize mannitol and phenylacetate but not glucose or arabinose as carbon sources. Sensitive to mezlocillin, cefamandole, cefotaxime, tetracycline, amikacin, and imipenem. Good growth and phenol biodegradation occur at 1–25°C (facultative psychrophile). Fully degrades up to 10 mM phenol as the sole carbon source. No growth occurs at pH 5 or 11. The predominant fatty acid (72.1%) is C15:0 anteiso, while C17:0 anteiso, C16:0 iso, C15:0 iso, C16:0, and C14:0 iso are detected in small amounts. The peptidoglycan type is A4α L-Lys–L-Glu. MK-10 is the predominant menaquinone, while MK-9 and MK-11 occur in smaller amounts (ratio of peak areas 72:12:1, respectively). The polar lipids are phosphatidylglycerol, diphosphatidylglycerol, phosphatidylinositol, and an unidentified glycolipid. The only cell-wall sugar is glucose.
Source: an alpine ice cave in Werfen (Salzburg), Austria.
Type strain: AG31, DSM 15454, JCM 13568, LMG 21914.
Sequence accession no. (16S rRNA gene): AJ616763.
Arthrobacter ramosus
Jensen 1960, 131AL
ra.mo'sus. L. masc. adj. ramosus branched, branching.
Motile by a few lateral flagella. Colonies on yeast extractpeptone media show no distinctive pigmentation. Nutritionally nonexacting: growth occurs in a suitable mineral salts medium with an ammonium salt or nitrate as sole nitrogen source. Starch is not hydrolyzed, and nicotine is not utilized.
The cell-wall peptidoglycan is of the Lys–Ala4 type (A3α). The whole-cell sugars are galactose, rhamnose, and mannose. The principal isoprenoid quinone is MK-9(H2) (Keddie et al., 1986) Major fatty acids are C15:0 anteiso (63%), C17:0 anteiso (8%), and C15:0 iso (6%) (Funke et al. 1996).
Source: beech forest soil at depth of 30–35 cm.
DNA G+C content (mol%): 62.2 (Tm).
Type strain: ATCC 13727, CIP 102361, DSM 20546, NBRC12958, JCM 1334, LMG 17309, NRRL B-3159, VKM Ac-1117.
Sequence accession no. (16S rRNA gene): X80742.
Arthrobacter rhombi
Osorio, Barja, Hutson and Collins 1999, 1220VP
rhom'bi. L. masc. n. rhombus flatfish; L. gen. n. rhombi of flatfish.
Cells are Gram-stain-positive, short rods and cocci. They are non-spore-forming and nonmotile. On BHA, smooth convex colonies which are yellowish or whitish in color and with a diameter of approximately 1 mm are formed after 48 h. Growth occurs in 1% (w/v) and 10% (w/v) NaCl and at 4 and 30°C. They are strictly aerobic and catalase- and oxidase-positive. Using the API CORYNE system, positive reactions for β-galactosidase and esculinase are observed. Negative reactions are obtained for nitrate reductase, pyrazinamidase, pyrrolidonyl arylamidase, urease, β-glucuronidase, N-acetyl-β-glucosaminidase, and gelatinase. Using the API 50CH system, the following substrates are used as sole carbon sources: glycerol, galactose, D-glucose, D-fructose, D-mannose, mannitol, amygdalin, arbutin, esculin, salicin, cellobiose, maltose, lactose, melibiose, sucrose, trehalose, β-gentiobiose, D-turanose, gluconate, and D-arabitol. Erythritol, D-arabinose, L-arabinose, ribose, D-xylose, adonitol, methyl-β-xyloside, L-sorbose, dulcitol, sorbitol, methyl-α-D-mannoside, methyl-α-D-glucoside, inositol, N-acetyl-β-glucosamine, inulin, melezitose, D-raffinose, starch, glycogen, xylitol, D-lyxose, D-tagatose, D-fucose, L-fucose, L-arabitol, 2-ketogluconate, and 5-ketogluconate are not utilized. The cell-wall murein type is A4α (L-Lys–L-Ala–D-Glu).
Source: organs of Greenland halibut (Reinhardtius hippoglossoides) in the deep waters of the Flemish Cap fishing ground (Newfoundland, NW Atlantic).
DNA G+C content (mol%): 61.0 (Tm).
Type strain: F.98.3HR.69, CCUG 38813, JCM 11678.
Other strains of the species: CCUG 38812.
Sequence accession no. (16S rRNA gene; type strain): Y15884.
Sequence accession no. (16S rRNA gene; another strain): Y15885.
Arthrobacter roseus
Reddy, Prakash, Matsumoto, Stackebrandt and Shivaji 2002, 1020VP
ro'se.us. L. masc. adj. roseus rose-colored.
Rod–coccus growth cycle with irregular rods in the exponential phase and predominantly coccoid cells (~1.0 µm in diameter) in the stationary phase. Gram-stain-positive, nonspore-forming, and nonmotile; colonies on peptone/yeast extract medium are round, smooth, convex, 0.2–1 mm in diameter, and red-pigmented. Pigment is insoluble in water but soluble in methanol and exhibits four absorption maxima, at 437, 467, 494, and 524 nm. Aerobic. Growth occurs at 5–30°C and at pH 6–12, and cells tolerate up to 5.8% NaCl. Optimum growth is observed at 22°C and pH 7. Catalase, phosphatase, gelatinase, and nitrate reduction tests are positive; oxidase, urease, lipase, β-galactosidase, arginine dihydrolase, indole production, methyl red test, Voges–Proskauer test, and levan formation are negative. Esculin and starch are not hydrolyzed. Positive for utilization of mannose, galactose, maltose, fructose, glucose, arabinose, ribose, xylose, rhamnose, raffinose, trehalose, succinic acid, fumaric acid, citric acid, mannitol, sorbitol, adonitol, sucrose, inulin, pyruvate, acetate, alanine, leucine, isoleucine, valine, serine, arginine, aspartic acid, glutamic acid, asparagine, glutamine, proline, and phenylalanine. The following substrates are not utilized as sole carbon sources: dulcitol, inositol, melibiose, lactose, nicotine, cellulose, glycine, threonine, cysteine, methionine, lysine, tyrosine, histidine, and tryptophan. No acid or gas from glucose, arabinose, xylose, rhamnose, fructose, galactose, mannose, lactose, maltose, or sucrose. Sensitive to penicillin, chlortetracycline, chloramphenicol, oxytetracycline, tetracycline, erythromycin, nitrofurantoin, bacitracin, nitrofurazone, leucomycin, rifampin, nystatin, cotrimoxazole, trimethoprim, ampicillin, carbenicillin, gentamicin, amoxicillin, tobramycin, and polymyxin B but resistant to furazolidone, colistin, furoxone, and kanamycin. The peptidoglycan type is Lys–Gly–Ala3 (variation A3α) and the major menaquinone is MK-9(H2). Cell-wall sugars are galactose, glucose, ribose, and rhamnose. Cellular fatty acids are C14:0 iso, C14:0, C15:0 iso, C15:0 anteiso, C15:0, C16:0 iso, C16:0, C16:1, C17:0 anteiso, C18:0, and C18:2. The predominant polar lipids are phosphatidylglycerol, diphosphatidylglycerol, phosphatidylinositol, and an unidentified glycolipid (Busse, unpublished results).
Source: a cyanobacterial mat sample from McMurdo Dry Valleys, Antarctica (77° 32′ 18” S, 160° 45′ E).
DNA G+C content (mol%): 66.0–69.0 (HPLC).
Type strain: CMS 90r, DSM 14508, JCM 11881, MTCC 3712.
Sequence accession no. (16S rRNA gene): AJ278870.
Arthrobacter russicus
Li, Kawamura, Fujiwara, Naka, Liu, Huang, Kobayashi and Ezaki 2004c, 834VP
rus'si.cus. N.L. masc. adj. russicus pertaining to Russia (Russian space station).
Grows well under aerobic conditions at 30°C on BHI agar plates, but unable to grow at 37°C. Gram-stain-positive, nonmotile, irregular rods, about 1.3–3.6 µm long and 0.5–0.9 µm wide. Circular, smooth and creamy colonies grow to a diameter of about 1.5 mm after 24 h. Positive for Tween 40, Tween 80, D-fructose, D-mannose, methyl pyruvate, L-alanyl glycine, and putrescine but negative for dextrin, arbutin, D-gluconic acid, maltose, maltotriose, D-melezitose, 3-methyl glucose, methyl-α-D-glucoside, methyl-β-D-glucoside, palatinose, D-ribose, salicin, sucrose, D-trehalose, acetic acid, α-hydroxybutyric acid, β-hydroxybutyric acid, γ-hydroxybutyric acid, α-ketovaleric acid, D-lactic acid methyl ester, L-lactic acid, L-malic acid, propionic acid, succinamic acid, succinic acid, alaninamide, L-asparagine, and 2,3-butanediol, esculin hydrolysis, urease activity, and nitrate reduction. Utilizes glucose but not inositol, sorbitol, lactose, xylitol, xylose, maltose, mannitol, and sucrose. Weakly positive for monomethyl succinate and L-glutamic acid. Variable for turanose, pyruvic acid, uridine, uridine 5′ monophosphate. Cell-wall peptidoglycan is A3α Lys–Ala2. Predominant isoprenoid quinone is MK-9(H2). Major cellular fatty acids are C15:0 anteiso and C17:0 anteiso. Predominant polar lipids are cardiolipin and phosphatidylinositol.
Source: an air sample from the Russian space station Mir.
DNA G+C content (mol%): 65.5 (HPLC).
Type strain: DSM 14555, GTC 863, JCM 11414.
Sequence accession no. (16S rRNA gene): AB071950.
Arthrobacter sanguinis
Mages, Frodl, Bernard and Funke 2008, 2985VP
san'gui.nis. L. masc. gen. n. sanguinis of blood, indicating that the bacterium was isolated from a blood culture.
The cells are coryneform bacteria without irregular branching, and spores are not formed. The organism is obligately aerobic. The colonies are whitish-grayish, slightly convex, of creamy texture, and up to 2 mm in diameter after 24 h of incubation at 35°C on Columbia SBA plates. Activities of the following enzymes are detected: catalase, acid phosphatase, alkaline phosphatase, esterase (C4), esterase lipase (C8), α-galactosidase, β-galactosidase, gelatinase, N-acetyl-β-glucosaminidase, α-glucosidase, leucine arylamidase, α-mannosidase, pyrazinamidase, and trypsin. Activities of α-chymotrypsin, cystine arylamidase, α-fucosidase, β-glucosidase, β-glucuronidase, lipase (C14), nitrate reductase, naphthol-AS-BI-phosphohydrolase, urease, and valine arylamidase are not observed. The bacterium is capable of utilizing N-acetylglucosamine, amygdalin, D-arabitol, cellobiose, fructose, galactose, gentiobiose, glucose, glycerol, maltose, mannitol, mannose, melibiose, potassium, gluconate, potassium 2-ketogluconate, raffinose, sucrose, sorbitol, trehalose, and turanose as carbon sources. The type strain did not utilize adonitol, D-arabinose, L-arabinose, L-arabitol, arbutin, dulcitol, erythritol, fucose, methyl-α-D-glucopyranoside, glycogen, inositol, inulin, potassium 5-ketogluconate, lactose, lyxose, methyl-α-D-mannopyranoside, melezitose, rhamnose, ribose, salicin, sorbose, starch, tagatose, methyl-β-D-xylopyranoside, xylitol, or xylose. Lysine is the diamino acid of the peptidoglycan, and C15:0 anteiso and C17:0 anteiso are the predominant cellular fatty acids.
Source: a blood culture.
DNA G+C content (mol%): not determined.
Type strain: CCUG 46407, DSM 21259.
Sequence accession no. (16S rRNA gene): EU086805.
Arthrobacter scleromae
Huang, Zhao, He, Wang, Liu, You and Guan 2005a, 1743VP (Effective publication: Huang, Zhao, He, Wang, Liu, You and Guan 2005b, 1453.)
scle.ro'ma.e. N.L. gen. n. scleromae of scleroma.
Cells are Gram-stain-positive, non-spore-forming, and nonmotile and display a rod–coccus life cycle. They are obligately aerobic, catalase-positive, and ~0.25 to ~0.35 µm in diameter. Colonies on blood agar or nutrient agar are whitish, glistening, convex, smooth surfaced, and circular. The colonies grow to up to 4 to 5 mm in size by 72 h. Growth occurs with a suitable carbon source in mineral salts medium; no additional growth factors are needed. Growth also occurs in the presence of 5% NaCl, at 15–37°C and pH 6–9, but not in 10% NaCl at 5 or 42°C. The organism hydrolyzes casein, DNA, esculin, gelatin, starch, and tyrosine but not lecithin or xanthine. The nitrate reduction test is weakly positive. N-Acetylglucosaminidase, β-galactosidase, α-glucosidase, lipase, pyrrolidonyl peptidase, and urease are not produced. Acid is produced from mannitol. It utilizes the following substrates as sole carbon sources: γ-aminobutyrate, L-arginine, L-asparagine, citrate, D-fructose, fumarate, D-galactose, D-gluconate, D-glucose, glycerol, L-histidine, p-hydroxybenzoate, 2-oxoglutarate, lactose, DL-malate, maltose, mannose, D-melezitose, phenylacetate, pyruvate, D-raffinose, salicin, sorbitol, succinate, sucrose, trehalose, D-turanose, xylitol, and D-xylose. The following substrates are not utilized: acetamide, adipate, adonitol, L-alanine, ρ-aminobenzoate, D-arabitol, arbutin, azelate, benzoate, 2,3-butanediol, n-butyrate, caprylate, D-cellobiose, L-citrulline, L-cysteine, dulcitol, erythritol, formate, glucosamine, D-glucuronate, glutarate, glycogen, glycolate, γ-hydroxybutyrate, inulin, DL-isoleucine, isovalerate, L-leucine, maleate, malonate, DL-methionine, α-methyl-D-glucoside, α-methyl-D-mannoside, nicotine, L-ornithine, oxalate, L-phenylalanine, o-phthalate, pimelate, L-rhamnose, D-ribose, sebacate, sorbose, suberate, D-tartrate, L-threonine, L-tryptophan, or DL-valine. Compounds slowly utilized are acetate, L-arabinose, inositol, mannitol, and melibiose. The strain is susceptible to ceftriaxone, chloramphenicol, rifampin, and tetracycline; moderately susceptible to cefazolin, cefotaxime, doxycycline, erythromycin, nitrofurantoin, piperacillin, and vancomycin; and resistant to amikacin, ampicillin, gentamicin, kanamycin, norfloxacin, oxacillin, penicillin G, streptomycin, and tobramycin. The major cellular fatty acid is C15:0 anteiso, with significant amounts of C15:0 iso, C16:0 iso, C17:1 anteiso ω9c, and C17:0 anteiso. The predominant menaquinone is MK-8(H2). The cell-wall peptidoglycan type is L-Lys–L-Ser–L-Thr–L-Ala (A3α), and the cell-wall sugars are galactose and glucose.
Source: swollen scleromata of a dermatosis patient.
DNA G+C content (mol%): 64.7 (Tm).
Type strain: YH-2001, CGMCC 1.3601, JCM 12642.
Sequence accession no. (16S rRNA gene): AF330692.
Arthrobacter soli
Roh, Sung, Nam, Chang, Kim, Yoon, Jeon, Oh and Bae 2008a, 1993VP (Effective publication: Roh, Sung, Nam, Chang, Kim, Yoon, Jeon, Oh and Bae 2008b, 43.)
so'li. L. neut. gen. n. soli of soil, the source of the type strain.
The cells are rod-shaped (0.9 × 2.0~3.0 µm), Gram-stain-positive, oxidase-positive, and catalase-negative. Colonies are yellow-pigmented and circular, measuring approximately 1.0–2.0 mm in diameter after 2 d of growth on TSBA at 30°C. The temperature range for growth is 16~40°C, but no growth occurs at 15 and 41°C. No growth occurs in NaCl at concentrations greater than 15%. Optimal pH for growth is 7.0. The strain can reduce nitrate to nitrite. Indole is not produced, and glucose fermentation does not occur. Cells are arginine dihydrolase- and urease-negative. Gelatin hydrolysis occurs, but not esculin and PNPG (p-nitrophenyl α-D-glucopyranoside) hydrolysis. Potassium gluconate, malate, trisodium citrate, phenylacetic acid, glycerol, L-arabinose, D-ribose, D-adonitol, methyl-β-D-xyloside, D-galactose, D-glucose, D-fructose, D-mannose, N-acetylglucosamine, D-cellobiose, D-maltose, D-lactose, gluconate, 2-ketogluconate, and 5-ketogluconate are assimilated; capric acid, adipic acid, erythritol, D-arabinose, D-xylose, L-xylose, L-sorbose, L-rhamnose, dulcitol, inositol, D-mannitol, D-sorbitol, methyl-α-D-mannoside, methyl-α-D-glucoside, amygdalin, arbutin, esculin, salicin, D-melibiose, sucrose, D-trehalose, inulin, D-melezitose, D-raffinose, starch, glycogen, xylitol, gentiobiose, D-turanose, D-lyxose, D-tagatose, D-fucose, L-fucose, D-arabitol, and L-arabitol are not assimilated. Cells are positive for alkaline phosphatase, esterase (C4), esterase lipase (C8), lipase (C14), leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, acid phosphatase, naphthol-AS-BI-phosphohydrolase, α-glucosidase, and α-mannosidase. However, cells are negative for α-chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, β-glucosidase, N-acetyl-β-glucosaminidase, and α-fucosidase. The predominant fatty acids are C15:0 anteiso and C17:0 anteiso, C16:0 iso, and C15:0 iso.
Source: wastewater reservoir sediment collected in Daejeon, Republic of Korea.
DNA G+C content (mol%): 62.0 (HPLC).
Type strain: SYB2, DSM 19449, KCTC 19291.
Sequence accession no. (16S rRNA gene): EF660748.
Arthrobacter stackebrandtii
Tvrzová, Schumann, Spröer, Sedláček, Verbarg, Kroppenstedt and Páčová 2005b, 807VP
sta.cke.brand'ti.i. N.L. gen. masc. n. stackebrandtii of Stackebrandt, named in honor of Erko Stackebrandt for his pioneering contributions to our insight into the phylogenetic structure of the suborder Micrococcineae and of the genus Arthrobacter in particular.
Cells are irregular club-shaped rods showing a rod–coccus cycle, 0.6–1 × 1–3 µm, occurring in pairs as typical V-forms. Gram-stain-positive, nonmotile, non-acid-fast, and nonspore-forming. Growth occurs at 4–30°C. Growth occurs at pH 5.7–9.1 and in the presence of 5% NaCl. Obligately aerobic. Positive for catalase, urease, and pyrazinamidase. Negative for oxidase, β-glucuronidase, β-galactosidase, α-glucosidase, N-acetyl-β-glucosaminidase, and pyrrolidonyl arylamidase. Nitrate is reduced to nitrite. Nitrite is not reduced; Tween 80 and esculin are not hydrolyzed. Starch and gelatin are hydrolyzed. Acid production is negative from glucose, ribose, xylose, mannitol, maltose, lactose, sucrose, and glycogen. The following compounds are utilized (Biolog system): α-cyclodextrin, dextrin, Tween 40, L-arabinose, D-arabitol, arbutin, D-cellobiose, D-fructose, L-fucose, D-galactose, α-D-glucose, α-D-lactose, lactulose, maltose, maltotriose, D-mannose, D-melibiose, methyl-α-D-galactoside, methyl-β-D-galactoside, palatinose, D-psicose, D-raffinose, D-ribose, salicin, stachyose, sucrose, D-trehalose, turanose, D-xylose, β-ketovaleric acid, methyl pyruvate, pyruvic acid, L-asparagine, glycerol, adenosine, inosine, thymidine, and uridine. Negative reactions (Biolog) were observed with β-cyclodextrin, glycogen, inulin, mannan, Tween 80, N-acetyl-D-glucosamine, N-acetyl-D-mannosamine, amygdalin, D-galacturonic acid, gentiobiose, D-gluconic acid, meso-inositol, D-melezitose, 3-methyl glucose, methyl-α-D-glucoside, methyl-β-D-glucoside, methyl-α-D-mannoside, L-rhamnose, sedoheptulosan, D-sorbitol, xylitol, β-hydroxybutyric acid, γ-hydroxybutyric acid, p-hydroxyphenylacetic acid, α-ketoglutaric acid, lactamide, D-lactic acid methyl ester, D-malic acid, monomethyl succinate, propionic acid, succinamic acid, succinic acid, N-acetylglutamic acid, alaninamide, D-alanine, L-alanine, L-alanyl glycine, L-glutamic acid, glycyl L-glutamic acid, L-pyroglutamic acid, putrescine, 2,3-butanediol, adenosine 5′-monophosphate, uridine 5′-monophosphate, fructose 6-phosphate, glucose 1-phosphate, glucose 6-phosphate, and DL-glycerophosphate. The cellular fatty acid contents of C15:0 anteiso (48.1%), C17:0 anteiso (14.4%), C14:0 iso (1.84%), C15:0 iso (14.2%), C16:0 iso (16.0%), C17:0 iso (3.5%), and C16:0 (1.4%). The peptidoglycan type is A3α Lys–Ala2. Strain CCM 2783T possesses predominantly menaquinone MK-9(H2) (62%) and MK-10(H2) (25%) and a small amount of MK-11(H2) (5%).
Source: poultry litter.
Type strain: CCM 2783, DSM 16005, JCM 14116.
Sequence accession no. (16S rRNA gene): AJ640198.
Arthrobacter subterraneus
Chang, Bae, Nam, Kwon, Park, Shin, Kim, Quan, Rhee, An and Park, 2008, 1993VP (Effective publication: Chang, Bae, Nam, Kwon, Park, Shin, Kim, Quan, Rhee, An and Park 2007, 1878.)
sub.ter.ra'ne.us. L. masc. adj. subterraneus under the earth, indicating the source of isolation.
The cells are Gram-stain-positive, short rods, and cocci (diameter 0.8–1 µm) occurring singly, in pairs, or in clusters. They are nonmotile and do not form endospores. Colonies grown on NA after 48 h are small (<1 mm), pale yellow, round with entire margins, of a low convexity, opaque, and smooth. Do not grow in an anaerobic chamber on NA. Optimum temperature for growth is 20–30°C. Weak growth at 37°C, and no growth at 45°C. Growth is apparent at 4°C after 1 week of incubation. Growth occurs on media with 13% NaCl, yet not with 15% NaCl. Grows at pH 5.3–10.5. Catalase-positive and oxidase-negative. Using the API STAPH system, positive reactions are observed for fermentation with D-mannitol, xylitol, D-melibiose, and raffinose, whereas negative reactions are obtained for fermentation with D-glucose, D-fructose, D-mannose, maltose, lactose, D-trehalose, potassium nitrate, β-naphthyl-acid phosphatase, sodium pyruvate, xylose, sucrose, α-methyl-D-glucoside, N-acetylglucosamine, arginine, and urea. Using the API ZYM system, activity is detected with esterase (C4), esterase lipase (C8), leucine arylamidase, valine arylamidase, trypsin, acid phosphatase, naphthol-AS-BI-phosphohydrolase, β-galactosidase, and α-glucosidase, whereas no activity is detected with alkaline phosphatase, lipase (14), cystine arylamidase, α-chymotrypsin, α-galactosidase, β-glucuronidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase. The predominant fatty acids are C15:0 anteiso (35%) and C17:1 anteiso ω9c (15%). The cell-wall peptidoglycan type is A3α, with a Lys–Thr–Ala3 interpeptide bridge. The major menaquinone is MK-9(H2). The cell-wall sugars are galactose, glucose, and ribose.
Source: deep subsurface water in Pohang, Gyeongbuk Province, Korea.
Type strain: CH7, DSM 17585, KCTC 9997.
Sequence accession no. (16S rRNA gene): DQ097525.
Arthrobacter sulfonivorans
Borodina, Kelly, Schumann, Rainey, Ward-Rainey and Wood 2002a, 685VP (Effective publication: Borodina, Kelly, Schumann, Rainey, Ward-Rainey and Wood 2002b, 180.)
sul.fo.ni.vo'rans. N.L. n. sulfonum sulfone group, L. v. vorare to devour, N.L. part. adj. sulfonivorans sulfone-devouring.
Cells are spherical, 0.7 µm in diameter. Gram-stain-positive, forming clumps and chains in liquid culture; motile; spores or capsules not seen; catalase- and oxidase-positive. Grows heterotrophically and aerobically on glucose, fructose, sucrose, galactose, acetate, ethanol, pyruvate, malate, succinate, citrate, serine, alanine, taurine, diethylsulfone, and yeast extract. Grows aerobically on methylated sulfur compounds (dimethylsulfone, dimethylsulfoxide, and dimethylsulfide), methanol, methylamine, and dimethylamine. Methylotrophic growth with dimethylsulfone uses the ribulose monophosphate cycle for formaldehyde assimilation. Does not grow with trimethylamine or alkanesulfonates (methane-, propane-, butane-, pentane-, and hexane-sulfonate), or autotrophically on inorganic sulfur compounds. Nitrate is not reduced. Growth on dimethylsulfone occurs optimally at pH 7.3–7.4 and at 20–25°C. Temperature range for growth is 4–30°C with no growth at 37 or 44°C. Ammonium chloride, nitrate, and methylamine are used as nitrogen sources for growth. Growth occurs in the presence of 1.5% or 2.5% (w/v) NaCl, but not with 5% (w/v) NaCl. On dimethylsulfone-agar medium, colonies are creamy-yellow, circular-umbonate, and 0.5–0.7 mm in diameter. The principal isoprenoid quinone is MK-9(H2), with smaller amounts of MK-10(H2), MK-8(H2), MK-9, MK-7(H2), and MK-11(H2). The principal cellular fatty acid is C15:0 anteiso, with C15:0 iso, C16:0 iso and C17:0 anteiso also present. Peptidoglycan contains lysine as the diagnostic diamino acid, as well as alanine, glutamic acid, threonine, and serine; peptidoglycan type (L-Lys–L-Ser–L-Thr–L-Ala) is A11.23.
Source: soil from the root ball of Allium aflatunense.
DNA G+C content (mol%): 61.0 (Tm).
Type strain: ALL, ATCC BAA-112, DSM 14002, JCM 13520.
Sequence accession no. (16S rRNA gene): AF235091.
Arthrobacter sulfureus
Stackebrandt, Fowler, Fiedler and Seiler 1984, 270VP (Effective publication: Stackebrandt, Fowler, Fiedler and Seiler 1983b, 484.)
sul.fu're.us. L. masc. adj. sulfureus of or like sulfur, meaning sulfur-colored.
In young cultures rod-shaped cells, irregular (0.4–0.6 × 1.2 to 1.6 µm), angular or slightly curved, and occasionally club-shaped; snapping divisions are observed. In old cultures, rods become shorter, and in some strains coccoid cells are predominant. Gram-stain-positive. Motile with peritrichous flagella or nonmotile. Colonies large, circular, smooth, entire, raised, glistening, dull, yellow, opaque, butyrous; 2–3 mm in diameter. Slightly turbid in nutrient broth; moderate growth and turbid in glutamate broth; liquefaction in nutrient gelatin stab culture. Nitrate not reduced to nitrite; indole not produced; hydrolysis of starch and casein negative; acetyl-methyl carbinol not produced; catalase-positive. No acid or gas from glycerol, xylose, glucose, sucrose, lactose, dextrin, and starch. Assimilate 4-hydroxybenzoate, L-leucine, L-asparagine, L-arginine, and L-histidine; two of three strains assimilate 5-aminovalerate, malonate, levulinate, D-ribose, D-galactose, D-xylitol, and 2,3-butylenglycol. Cellulose not attacked. The type strain is positive for pyrazinamidase and urease but negative for β-galactosidase. Assimilation of D-arabitol, inositol, maltose, mannitol, xylitol, and 5-ketogluconate, but no assimilation amygdalin, arbutin, cellobiose, D-mannose, D-turanose, D-xylose, glycerol, L-arabinose, melibiose, rhamnose, ribose, salicin, sucrose, trehalose, β-gentiobiose, and N-acetylglucosamine (only type strain investigated; Osorio et al., 1999). The peptide subunit consists of alanine, D-glutamic acid, and lysine. The interpeptide bridge consists of glutamic acid (Lys–Ala–Glu type, variation A4α). Cell wall (only one strain tested) contains galactose and traces of mannose. The long-chain fatty acids (two strains tested) are primarily of the iso, anteiso, straight chain, and unsaturated acid types, with the anteiso 12-methyltetradecanoic acid (C15:0 anteiso) dominating. The major respiratory quinones are unsaturated menaquinones either with 9 or with equal amounts of 9 and 10 isoprenoic units. Polar lipids consist of diphosphatidylglycerol, phosphatidylglycerol, and of unidentified glycolipids. Optimal temperature and pH for growth are 25–30°C and 7.0–8.0. No growth at pH 5.0.
Source: soil.
DNA G+C content (mol%): 64.5–66.0 (Tm).
Type strain: ATCC 19098, CIP 106986, DSM 20167, NBRC12678, JCM 1338, LMG 16694, NRRL B-14730.
Sequence accession no. (16S rRNA gene): X83409.
Sequence accession no. (recA): AF214787.
Arthrobacter tecti
Heyrman, Verbeeren, Schumann, Swings and De Vos 2005, 1462VP
tec'ti. L. gen. n. tecti from the ceiling.
Cells are Gram-stain-positive, short rods and cocci (diameter 0.8–1 µm) occurring in pairs or clusters. They are nonmotile and do not form endospores. Colonies on NA after 48 h are small (<1 mm), yellow, round with entire margins, of low convexity, opaque, and smooth. No growth in an anaerobic chamber on NA. Optimum temperature for growth is 22–30°C. Growth on medium with 15% NaCl added. Growth at pH 6–9; optimal growth at pH 8. Catalase-positive and oxidase-negative. Using the API CORYNE system, a positive reaction is observed for N-acetyl-β-glucosaminidase. Most strains, including the type strain, test positive for gelatinase but negative for pyrrolidonyl arylamidase and fermentation of ribose and mannitol. Negative reactions are obtained for nitrate reduction, β-glucuronidase, urease, and fermentation of ribose, xylose, maltose, lactose, and glycogen. Variable reactions are obtained for pyrazinamidase, alkaline phosphatase, β-galactosidase, α-glucosidase, esculin (β-glucosidase), and fermentation of glucose and sucrose. Using the API ZYM system, activity is detected for alkaline phosphatase (weak), acid phosphatase (weak), leucine arylamidase, and phosphoamidase (weak). Most strains test positive for esterase C4, esterase lipase C8, trypsin (negative for the type strain), N-acetyl-β-glucosaminidase (negative for the type strain), and α-mannosidase (negative for the type strain). No activity is detected for lipase C14, valine arylamidase, cystine arylamidase, α-galactosidase, β-glucuronidase, or α-fucosidase. Variable reactions are obtained for chymotrypsin, β-galactosidase, α-glucosidase, and β-glucosidase. Predominant fatty acids are C15:0 anteiso (44%) and C15:0 iso (37%). The peptidoglycan type is A3α Lys–Thr–Ala3. MK-9(H2) (41%), MK-10(H2) (28%), and MK-11(H2) (11%) are the predominant menaquinones, while MK-12(H2), MK-9, MK-10, and MK-11 occur in only small amounts. The cell-wall sugars are galactose and rhamnose. Polar lipids of the type strain are phosphatidylinositol, phosphatidylglycerol, diphosphatidylglycerol, and one unknown phospholipid.
Source: a biofilm overgrowing the ceiling of the main room of the Servilia tomb (Roman Necropolis of Carmona, Spain).
DNA G+C content (mol%): 63.7 (HPLC).
Type strain: DSM 16407, JCM 21772, LMG 22282.
Another strain: LMG 22285.
Sequence accession no. (16S rRNA gene; type strain): AJ639829.
Sequence accession no. (16S rRNA gene; another strain): AJ639828.
Arthrobacter tumbae
Heyrman, Verbeeren, Schumann, Swings and De Vos 2005, 1463VP
tum'ba.e. L. gen. n. tumbae of a tomb.
Cells are Gram-stain-positive, short rods and cocci (diameter, 0.8–1 µm) occurring singly, in pairs, or in clusters. They are nonmotile and do not form endospores. Colonies on NA after 48 h are small (<1 mm), yellow–orange, round with entire margins, of low convexity, opaque, and smooth. Do not grow in an anaerobic chamber on NA. Optimum temperature for growth is 22–30°C. Weak growth at 37°C, and no growth at 52°C. Growth at 4°C after 1 week of incubation. Growth on medium with 10% NaCl, but not with 15% NaCl. Catalase-positive and oxidase-negative. Alkaliphilic; pH range for growth of 7–10 with an optimum of 8–9. Using the API CORYNE system, positive reactions are observed for pyrazinamidase, β-glucuronidase, β-galactosidase, and α-glucosidase. Most strains, including the type strain, test positive for gelatinase. Negative reactions are obtained for N-acetyl-β-glucosaminidase and fermentation of ribose, xylose, mannitol, and glycogen. Variable reactions are obtained for nitrate reduction, pyrrolidonyl arylamidase, alkaline phosphatase, esculin (β-glucosidase), urease, and fermentation of glucose, maltose, lactose, and sucrose. Using the API ZYM system, activity is detected for esterase C4, leucine arylamidase, and phosphoamidase (weak). Most strains, including the type strain, test positive for esterase lipase C8 and negative for alkaline phosphatase, valine arylamidase, α-galactosidase, and β-galactosidase. No activity is detected for lipase C14, cystine arylamidase, β-glucuronidase, β-glucosidase, N-acetyl-β-glucosaminidase, or α-fucosidase. Variable reactions are obtained for trypsin, chymotrypsin, acid phosphatase, α-glucosidase, and α-mannosidase. Predominant fatty acids are C15:0 anteiso (57%) and C15:0 iso (17%). The peptidoglycan type is A3α Lys–Thr–Ala3. MK-9(H2) (54%) and MK-10(H2) (25%) are the predominant menaquinones, while MK-7(H2), MK-8(H2), and MK-11(H2) occur in only small amounts. The cell-wall sugar is galactose. Polar lipids of the type strain are phosphatidylinositol, phosphatidylglycerol, diphosphatidylglycerol, and one unknown phospholipid.
Source: a biofilm overgrowing a mural painting in the Servilia tomb (Roman Necropolis of Carmona, Spain).
DNA G+C content (mol%): 64.7 (HPLC).
Type strain: DSM 16406, JCM 21773, LMG 19501.
Sequence accession no. (16S rRNA gene): AJ315069.
Arthrobacter uratoxydans
Stackebrandt, Fowler, Fiedler and Seiler 1984, 270VP (Effective publication: Stackebrandt, Fowler, Fiedler and Seiler 1983b, 483.)
u.ra.to'xy.dans. N.L. n. uratum salt of uric acid; N.L. part. adj. oxydans oxidizing; N.L. part. adj. uratoxydans uric acid oxidizing.
In young cultures, straight to slightly curved or ellipsoidal rods with metachromic granules to pleomorphic rods which are club-shaped swellings; angular or palisade rods formed by snapping divisions are observed. Gram-stain-positive; 0.8–1.2 × 1.0–5 µm; nonmotile. No capsules; no endospores. Colonies circular, flat, smooth, entire margins, slightly glistening and opaque, 3–4 mm in diameter; creamy white in young cultures to pale yellow in old cultures. Water soluble, pale yellowish brown pigment produced. In nutrient broth, scanty surface growth; no sediments. In gelatin stab cultures, best growth at top and filiform; liquification stratiform. In litmus milk no reaction; slightly alkaline; no coagulation. Nitrate reduced to nitrite. Voges–Proskauer test negative. Indole production negative; hydrogen sulfide production positive; hydrolysis of starch negative; utilization of sodium citrate (Koser's and Christen's medium) positive; utilization of a great variety of organic nitrogen compounds. Uricase strongly positive; hemolysis negative; nonpathogenic. The type strain is positive for pyrazinamidase but negative for β-galactosidase. It can assimilate N-acetylglucosamine but not amygdalin, arbutin, cellobiose, D-arabitol, D-mannose, D-turanose, D-xylose, galactose, glycerol, inositol, L-arabinose, maltose, mannitol, melibiose, salicin, sucrose, trehalose, xylitol, β-gentiobiose, or 5-ketogluconate (only type strain investigated; Osorio et al., 1999). Utilizes 5-amino valerate, glyoxylate, L-leucine, L-xylose, and uric acid. Hydrolysis of xanthine and casein; acid is not produced from sugars; cellulose is not decomposed. Aerobic to slightly facultative anaerobic; optimal temperature and pH for growth, 27–35°C and 7–8; thermal death time 10 min at 52°C. The peptide subunit of the peptidoglycan consists alanine, D-glutamic acid, and lysine. The interpeptide bridge contains alanine and glutamic acid (Lys–Ala–Glu type, variation A4α). Major fatty acids are C15:0 anteiso (34%), C15:0 iso (27%), C17:0 anteiso (14%), C17:0 iso (11%) and C16:0 iso (7%) (Funke et al., 1996).
Source: humus soil.
DNA G+C content (mol%): 61.2–61.5 (Tm).
Type strain: ATCC 21749, CIP 102367, DSM 20647, NBRC 15515, JCM 11944, LMG 16220, VKM Ac-1979.
Sequence accession no. (16S rRNA gene): X83410.
Sequence accession no. (recA): AF214791.
Arthrobacter ureafaciens
(ex Krebs and Eggleston 1939) Clark 1955, 112AL (Corynebacterium ureafaciens Krebs and Eggleston 1939, 310)
u.re.a.fa'ci.ens. N.L. n. urea urea; L. v. facio to make, produce; N.L. part. adj. ureafaciens urea-producing.
Cells are nonmotile. Colonies on yeast extract-peptone media are pale gray, becoming yellow especially when incubated in diffuse daylight at ~20°C. When supplied with biotin, cells grow in suitable mineral salts medium with an ammonium salt as sole nitrogen source and with glucose as carbon and energy source (Keddie et al., 1986). Starch is not hydrolyzed, nitrate is not reduced, and cells do no grow in 10% NaCl. Nicotine blue is not produced from nicotine. Utilizes L-arginine, L-asparagine, L-arabinose, D-galactose, D-glucose, D-ribose, D-xylose, histidinol, inositol, 4-aminobutyrate, and ρ-hydroxybenzoate but not L-histidine, L-leucine, L-rhamnose, butanediol, or malonate. Assimilate citric acid, formic acid, glutaric acid, propionic acid, and uric acid but not adipic acid, benzoic acid, malonic acid, or pimelic acid. Urea is formed from creatinine and uric acid. The cell-wall peptidoglycan is of the Lys–Ala–Thr–Ala type. The whole-cell-wall sugar is galactose (mannose). The principal isoprenoid quinone is MK-9(H2). The predominant fatty acid is C15:0 anteiso (58%), followed by C17:0 anteiso (28%), C16:0 iso (6%), and C15:0 iso (5%) (Kodama et al., 1992).
Source: soil.
DNA G+C content (mol%): 61.7 (Tm).
Type strain: ATCC 7562, CIP 67.3, DSM 20126, NBRC 12140, JCM 1337, LMG 3812, VKM Ac-1121.
Sequence accession no. (16S rRNA gene): X80744.
Sequence accession no. (recA): AF214782.
Arthrobacter woluwensis
Funke, Hutson, Bernard, Pfyffer, Wauters and Collins 1997a, 242VP (Effective publication: Funke, Hutson, Bernard, Pfyffer, Wauters and Collins, 1996, 2362.)
wo.lu.wen'sis. N.L. masc. adj. woluwensis of or belonging to Woluwe, a town near Brussels, Belgium, where the type strain was isolated from a patient.
The cells are coryneform bacteria which develop into cocci when the cultures become older (72 h). Jointed rods are observed after 1–2 d of incubation. No spores formed. The cells are nonmotile. The organism is obligately aerobic. The colonies are whitish-grayish, smooth, convex, and larger than 2 mm in diameter after 24 h of incubation at 37°C in 5% CO2 on SBA. The organism is catalase-positive. Nitrate is not reduced. Urea (72 h) and esculin (24 h) are hydrolyzed. DNase and gelatinase activities are detected within 24 h. Glycerol, galactose, D-glucose, D-fructose, D-mannose, dulcitol, mannitol, sorbitol, N-acetylglucosamine, amygdalin, arbutin, salicin, cellobiose, maltose, lactose, melibiose, sucrose, trehalose, melezitose, D-raffinose, β-gentiobiose, D-turanose, D-arabitol, gluconate, 2-keto-gluconate, and 5-keto-gluconate are utilized. Erythritol, D-arabinose, L-arabinose, ribose, D-xylose, L-xylose, adonitol, β-methyl-xyloside, L-sorbose, rhamnose, inositol, α-methyl-D-mannoside, α-methyl-D-glucoside, inulin, starch, glycogen, xylitol, D-lyxose, D-tagatose, D-fucose, L-fucose, and L-arabitol are not utilized. The following enzyme activities are detected: alkaline and acid phosphatase, esterase (C4), esterase lipase (C8), lipase (C14), leucine arylamidase, cystine arylamidase, trypsin, β-galactosidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, and α-mannosidase. Chymotrypsin, α-galactosidase, β-glucuronidase, and α-fucosidase are not present. The peptidoglycan type is A4α, L-lysine–D-aspartic acid. Major fatty acids are C15:0 anteiso (38%), C15:0 iso (22%), C17:0 anteiso (15%), C17:0 iso (6%), and C16:0 iso (11%) (Funke et al., 1996).
Source: cultures of human blood.
DNA G+C content (mol%): 69.0 (HPLC).
Type strain: CUL 1808, ATCC 700220, CCUG 36790, CIP 104908, DSM 10495, JCM 11679.
Sequence accession no. (16S rRNA gene): X93353.
Misclassified species
-
Arthrobacter viscosus Gasdorf, Benedict, Cadmus, Anderson and Jackson 1965, 150AL
vis.co'sus. L. masc. adj. viscosus viscous, sticky; referring to exopolysaccharide production and viscous colonies.
Arthrobacter viscosus was the name given to two polysaccharide-producing bacterial strains. Both strains are considered to be Gram-stain-negative, motile, aerobic rods (ATCC catalog, 1982). The type strain produces from glucose large quantities of a carbohydrate polymer based on galactose, glucose, and mannuronic acid, and contains meso-diaminopimelic acid (meso-DAP) as cell-wall diamino acid but neither arabinose nor mycolic acids (Keddie and Cure, 1978). The second strain of the species, ATCC 19583, also contains a directly linked peptidiglycan based on meso-DAP and lacks arabinose in the cell wall (Schleifer and Kandler, 1972).
Source: Guatemalan soil.
DNA G+C content (mol%): 59.4 (method of analysis specified in Skyring et al., 1971).
Type strain: ATCC 19584; CIP 82.105; DSM 7307; JCM 11566; LMG 16473; NCIB 9729; NRRL B-1973.
Sequence accession no. (16S rRNA gene): AJ639832.



