Abstract
Co.ry.ne.bac.te'ri.um. Gr. n. coryne a club; L. neut. n. bacterium a rod, and in biology a bacterium (so called because the first ones observed were rod-shaped); N.L. neut. n. Corynebacterium a club bacterium.
Actinobacteria / Actinobacteria / Corynebacteriales / Corynebacteriaceae / Corynebacterium
Straight to slightly curved rods with tapered ends. Rods are usually short or of medium length. Club-shaped forms may be observed; sometimes ellipsoidal, ovoid or rarely, “whip handles” (see below, Corynebacterium matruchotii) or thinner rods with bulges (see below, Corynebacterium sundsvallense) observed. Snapping division produces angular and palisade arrangements of cells. Gram-stain-positive; some cells stain unevenly. Metachromatic (synonym being polyphosphate) granules may be observed for some species. Not-acid-fast (Ziehl–Neelsen stain), and no species has aerial mycelium. Nonsporeforming. All species are nonmotile. All species are catalase positive. All species are oxidase negative except for Corynebacterium bovis, Corynebacterium aurimucosum, Corynebacterium doosanense, and Corynebacterium maris (below). Many species are facultatively anaerobic and some are aerobic. Chemoorganotrophs. Some species are lipophilic. Many species produce acid from glucose and some other sugars in peptone media. Several species alkalinize citrate as sole carbon sources, but most do not.
DNA G+C content (mol%): 46–74.
Type species: Corynebacterium diphtheriae (Kruse 1886) Lehmann and Neumann 1896, 350 (“Bacillus diphtheria” Kruse in Flügge 1886, 225).
Straight to slightly curved rods with tapered ends. Rods are usually short or of medium length. Club-shaped forms may be observed; sometimes ellipsoidal, ovoid or rarely, “whip handles” (see below, Corynebacterium matruchotii) or thinner rods with bulges (see below, Corynebacterium sundsvallense) observed. Snapping division produces angular and palisade arrangements of cells. Gram-stain-positive; some cells stain unevenly. Metachromatic (synonym being polyphosphate) granules may be observed for some species. Not-acid-fast (Ziehl–Neelsen stain), and no species has aerial mycelium. Nonsporeforming. All species are nonmotile. All species are catalase positive. All species are oxidase negative except for Corynebacterium bovis, Corynebacterium aurimucosum, Corynebacterium doosanense, and Corynebacterium maris (below). Many species are facultatively anaerobic and some are aerobic. Chemoorganotrophs. Some species are lipophilic. Many species produce acid from glucose and some other sugars in peptone media. Several species alkalinize citrate as sole carbon sources, but most do not.
Cell-wall peptidoglycan is based on meso-diaminopimelic acid (meso-DAP) (variation of A1γ of Schleifer and Kandler). Glycan type of cell walls contains acetyl residues. Major cell-wall sugars are arabinose and galactose (also referred to as arabinogalactan), but occasionally other sugars detected. Short-chain mycolic acids (also referred to as corynomycolates, corynemycolates, or α-alkyl-β-hydroxy-long-chain fatty acids) 22–36 carbons in length may be present, but some species lack mycolates entirely (see below, Corynebacterium amycolatum, Corynebacterium atypicum, Corynebacterium caspium, Corynebacterium ciconiae, and Corynebacterium kroppenstedtii). Long-chained cellular fatty acids are of the straight-chain saturated and monounsaturated types, with significant amounts of hexadecanoic (palmitic, C16:0), octadecanoic (C18:0), and cis-9-octadecenoic (“oleic”, C18:1 ω9c) acids as major components. Small or moderate amounts of tuberculostearic acid (TBSA) (10-methyl C18:0) and other cellular fatty acids may also be present. Corynebacterium diphtheriae, Corynebacterium ulcerans, Corynebacterium pseudotuberculosis, and Corynebacterium vitaeruminis are the only species where significant amounts of the 16:1 isomer, C16:1 ω7c, are observed and corynemycolates may be observed to coelute with cellular fatty acids (Bernard et al., 1991). Branched-chain or hydroxylated fatty acids are absent or found only in trace amounts. Metabolic products of fermentation may include small volumes of acetic, succinic, and lactic acids, but production of propionic acid is species specific (Bernard et al., 2002). Dihydrogenated menaquinones with either eight [MK-8(H2)] and/or nine [MK-9(H2)] isoprene units are present. In addition, MK-7(H2) has been detected for Corynebacterium glaucum and Corynebacterium lubricantis; small amounts of MK-10(H2) have also been found for Corynebacterium thomssenii (below). Phospholipids include simple, phosphatidylinositol, phosphatidylinositol dimannoside(s), phosphatidylglycerol trehalose dimycolates, and other glycolipids (Collins and Cummins, 1986; Yague et al., 1997). Phosphatidyethanolamine is absent except for Corynebacterium bovis and Corynebacterium urealyticum (Kämpfer et al., 1999).
DNA G+C content (mol%): 46–74.
Type species: Corynebacterium diphtheriae (Kruse 1886) Lehmann and Neumann 1896, 350 (“Bacillus diphtheria” Kruse in Flügge 1886, 225).
Number of validated species: 84
Further descriptive information
The genus Corynebacterium contains 84 validly published named species at the time of writing (May 2010). Phylogenetic relatedness among Corynebacterium species was first inferred by nearly full 16S rRNA gene sequence relationship studies in 1995 (Pascual et al., 1995; Ruimy et al., 1995) (Figure 1). Since the 1st edition of the Manual, members of this genus have been restricted to species most closely related by 16S rRNA gene sequencing to the type species, Corynebacterium diphtheriae. The genus Corynebacterium and the closely related genus, Turicella, are sole genera in the family Corynebacteriaceae (Ludwig et al., 2009a; Zhi et al., 2009). In addition to a close relationship by 16S rRNA or rpoB gene sequence analyses (described below), Corynebacterium species share phenotypic, chemotaxonomic, and other commonalities, and so taxa from the 1st edition of this Manual which do not fit this description have now been reassigned to other genera and families (outlined below). The degree of variance using 16S rRNA gene sequencing is 2% or greater for many species in this genus, and so most are discernable from each other without study of additional gene targets if comparison of nearly full (∼1400–1500 bp) sequence of the 16S rRNA gene is used (Stackebrandt and Ebers, 2006). Shorter, but more rapidly analyzed lengths (400–500 bp) of 16S rRNA gene sequence have been used for characterization of Corynebacterium species for rapid identification (Tang et al., 2000). However, when full sequencing data for the 16S rRNA gene for all taxa are analyzed, some species are related with a degree of variance which is ≤2%. These include: Corynebacterium afermentans, Corynebacterium coyleae, Corynebacterium mucifaciens (<2%); Corynebacterium aurimucosum, Corynebacterium minutissimum, and Corynebacterium singulare (<2%); Corynebacterium sundsvallense and Corynebacterium thomssenii (<1.5%); Corynebacterium ulcerans and Corynebacterium pseudotuberculosis (<1% to each other, both <2% to Corynebacterium diphtheriae); Corynebacterium propinquum and Corynebacterium pseudodiphtheriticum (<2%); Corynebacterium xerosis, Corynebacterium freneyi, and Corynebacterium hansenii (<2%); and Corynebacterium macginleyi and Corynebacterium accolens (<2%).
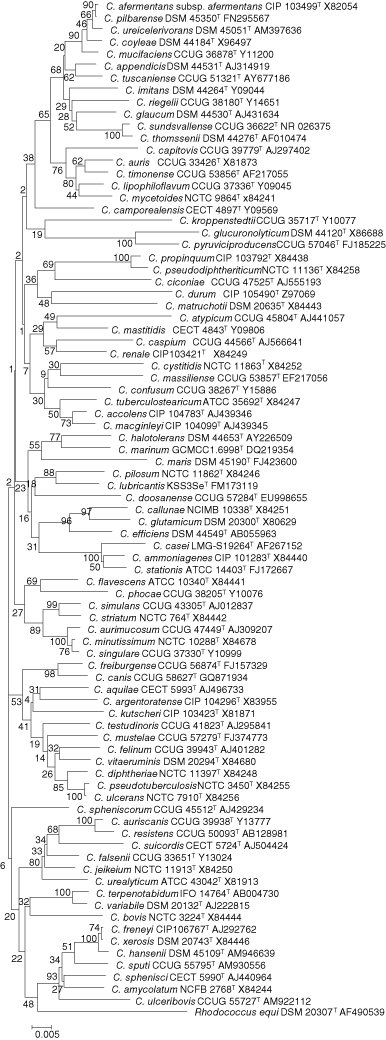
Corynebacterium species which cannot be discerned by 16S rRNA gene sequencing can be further characterized by sequencing of other gene targets. One widely used method is that of rpoB gene sequencing, as an adjunct to or in lieu of, 16S rRNA gene sequencing. When full or partial rpoB gene sequences were used, taxa with ≥95% identity are thought to be members of the same species and those with ≥98% identity were deemed to be members of the same subspecies (Khamis et al., 2004, 2005). Corynebacterium xerosis, Corynebacterium freneyi, and Corynebacterium hansenii, which could not be discerned using 16S rRNA, 16S–23S spacer region or rpoB gene sequencing, were determined only by DNA–DNA hybridization to be separate species (Renaud et al., 2007). Restriction length polymorphism using cfoI fragment of the 16S–23S region has been used to characterize Corynebacterium freneyi strains (Funke and Frodl, 2008b). Sequencing of a gene associated with cell division, divIVA, has been applied to discern Corynebacterium amycolatum from closely related species (Letek et al., 2006). A PCR-based assay to detect dtxR, a chromosomal iron-dependent repressor gene associated with Corynebacterium diphtheriae strains, has been used as a method to screen for these organisms (Pimenta et al., 2008). A number of other approaches have been used to try to separate closely related Corynebacterium species. Amplified rRNA-restriction analysis (ARDRA) using the enzymes aluI, cfoI, and rsaI was applied as a means to differentiate among otherwise closely related species (Vaneechoutte et al., 1995). Length polymorphisms of the 16S–23S rRNA gene spacer region were used to reveal unexpected heterogeneity among strains of species otherwise thought to be monophyletic (Aubel et al., 1997). Ribotyping and RFLP (restriction fragment length polymorphisms) have been used to differentiate species using various enzymes (Björkroth et al., 1999). Entire genome studies of Corynebacterium species have been completed to date for Corynebacterium glutamicum (ATCC 13032T) (Kalinowski et al., 2003), Corynebacterium efficiens (YS-314T) (Nishio et al., 2003), Corynebacterium diphtheriae (NCTC 13129) (Cerdeno-Tarraga et al., 2003), Corynebacterium jeikeium (strain K411) (Tauch et al., 2005), Corynebacterium urealyticum (DSM 7109T) (Tauch et al., 2008b), Corynebacterium kroppenstedtii (DSM 44385T) (Tauch et al., 2008a), and a black-pigmented Corynebacterium aurimucosum strain (CN-1) (Trost et al., 2010). These range in size from Corynebacterium urealyticum (2,369,217 bp) to Corynebacterium glutamicum (3,309,401 bp), with a number of additional projects involving Corynebacterium species under way as of this writing. Although still in its infancy, comparative analysis of these entire genomes has provided insight into common or different protein expressions found among these taxa including a set of conserved, DNA-binding transcriptional regulators consisting of 28 proteins that is involved with the regulation of cell division, septation, SOS and stress response, carbohydrate metabolism, and macroelement and metal homeostasis (Brune et al., 2005).
Corynebacterium species epidemiologically linked in an outbreak have been characterized comprehensively using several molecular typing schemes. Concerted efforts to track a large diphtheria outbreak in former Soviet Union (FSU) states, spearheaded by the European laboratory working group on diphtheria (DIPNET), have been studied by various methods, particularly rRNA gene restriction pattern determination (ribotyping). Multilocus enzyme electrophoresis (MEE), yielding an electromorph (ET) type, pulsed field gel electrophoresis (PFGE), randomly amplified polymorphic DNA (RAPD), and amplified fragment length polymorphism (AFLP) analyses have been used to characterize outbreak strains (De Zoysa et al., 1995b; Popovic et al., 1996) and isolates which predated the outbreak from those geographic areas (Skogen et al., 2002). By these means, a clonal group of closely related strains, particularly ribotypes Sankt-Peterburg, Rossija and others, were found to be largely responsible for the Russian and FSU state countries outbreak. With this information, movement of clonal strains to other countries could be tracked (Mokrousov et al., 2005). Four molecular methods (ribotyping, AFLP, PFGE, and RAPD) for typing Corynebacterium diphtheriae strains have been compared. Ribotyping was found to provide the most useful and discriminatory information (De Zoysa et al., 2008) and used to propose international nomenclature rules (Grimont et al., 2004). A multi-method approach was used to characterize a large number of American and Canadian isolates; clonal groups distinct from European outbreak strains and in circulation for many years, in spite of universal vaccination programs in both countries, were found to persist (Marston et al., 2001). Further discrimination among strains recovered during the Russian/FSU state outbreak have been studied using a microarray approach, the use of which could arguably eliminate inherent laboratory inter-reproducibility issues or cumbersome methods particularly associated with PFGE, MEE, and ribotyping (Mokrousov et al., 2009). A multilocus, sequence typing (MLST) scheme has been described for Corynebacterium diphtheriae surveillance and data derived from sequencing seven housekeeping genes (atpA, dnaE, dnaK, fusA, leuA, odhA, rpoB) were curated, compared to a database, assigned a sequence type, and interpreted (Bolt et al., 2010; Dallman et al., 2008).
Molecular typing methods for Corynebacterium species other than Corynebacterium diphtheriae have been occasionally described. Thirty-two strains of Corynebacterium urealyticum from humans or animals, recovered over several years, were extensively characterized by ribotyping. There, most human isolates were generally found to be multidrug-resistant (MDR) and clustered into ribotypes called 8, 9, and 10, in contrast with animal strains which were assigned to ribotypes 5 and 6 and were more susceptible to antibiotics (Nieto et al., 2000). A unique strain of Corynebacterium striatum which produced a diffusible brown pigment and was associated with respiratory, wound, or blood infections in an intensive care unit (ICU) was found to be clonal, when studied by DNA restriction fragment patterns and Southern hybridization with an att site probe, whereas apigmented, contemporary strains to the outbreak had diverse genetic patterns (Leonard et al., 1994). RAPD method was used to study an outbreak among patients in a surgical ICU, where a genetically and biochemically identical clone with identical antibiogram, had spread from patient to patient and could be discerned from other strains circulating in the ward over a 1 year period (Brandenburg et al., 1996). Ribotyping and RAPD were used to prove that cutaneous sites and blood culture isolates of Corynebacterium striatum from the same patient were identical (Martin et al., 2003). PFGE was used to evaluate 36 MDR Corynebacterium striatum strains recovered primarily from ventilatory-related respiratory or central venous catheter infections from three hospitals over a 2-year period, where infection was associated with a single clone possessing ermX, tet A/B, cmx A/B, and aphA1 resistance genes (Campanile et al., 2009). PFGE was used to type 48 Corynebacterium striatum strains recovered from patients in a community hospital over a 5-year period, primarily from respiratory specimens; these could be assigned into 14 patterns with 20 subtypes and provide definitive linkages for nosocomial acquisition (Otsuka et al., 2006). Strains of Corynebacterium ulcerans recovered from a patient and her dog were found to be identical but differed from reference strains when characterized by ribotyping (Lartigue et al., 2005). Ribotyping was also used to link infection by a toxigenic strain of Corynebacterium ulcerans with a patient and companion animals (two dogs, one cat) (Hogg et al., 2009). An outbreak of fluoroquinolone-resistant Corynebacterium macginleyii was studied by MLST of seven housekeeping genes (adk, dnaA, fumC, gltA, gyrB, icd, and purA) in parallel to RAPD analysis, with the two methods being found to be equally discriminating (Eguchi et al., 2008). Corynebacterium pseudotuberculosis outbreaks in sheep and goats have been studied using PFGE (Connor et al., 2007).
Metabolic pathways and metabolism
Corynebacterium species historically have been characterized as having metabolisms which are fermentative, oxidative, or neither fermentative or oxidative (von Graevenitz and Bernard, 2003). Three closely related species, Corynebacterium glutamicum, Corynebacterium callunae, and Corynebacterium efficiens, have had very extensive metabolic, chemotaxonomic, and genetic properties studied because of their usefulness in biotechnological production of amino acids (Bayan et al., 2003; Wendisch et al., 2006). Comprehensive metabolic pathways and putative functions have been conjectured for species where complete genome studies have been done (Brune et al., 2005; Cerdeno-Tarraga et al., 2003; Kalinowski et al., 2003; Mokrousov, 2009; Nishio et al., 2003; Nishio et al., 2004; Tauch et al., 2008a, 2008b; Wendisch et al., 2006).
Pathogenicity, ecology, miscellaneous
Corynebacterium diphtheriae, Corynebacterium ulcerans, and Corynebacterium. pseudotuberculosis are the only species which may produce diphtheria toxin (DT), a potent exotoxin which plays a significant role in pathogenicity. The resultant disease, diphtheria, has historically been associated with significant morbidity and mortality in humans and animals, prior to universal use of an efficacious vaccine in countries around the world with higher socio-economic means (Funke et al., 1997f). In diphtheria, DT contributes to the formation of a pseudomembrane in the nasopharynx of the patient; although the organism is rarely found outside the infected area, the toxin, once absorbed by the circulatory system, can cause systemic complications, such as myocarditis and neuritis. Therefore, early administration of diphtheria antitoxin (DAT), a commercial immunoglobulin which neutralizes circulating DT, is critical for patient care, but local stockpiles of DAT may not exist or be in limited supply in countries where diphtheria has low prevalence (Wagner et al., 2009). Diphtheria is still endemic in some subtropical and tropical countries as well as among individuals of certain ethnic groups (e.g. indigenous peoples in the Americas and Australia). At population level, endemic geographically-specific variants of Corynebacterium diphtheriae are influenced by human host factors, public health control efforts (i.e. universal vaccination), and socioeconomic conditions. Importation of (toxigenic) strains, or lysogenization by a bacteriophage (also called corynephage) of local strains, may instigate an outbreak (Mokrousov, 2009).
It has been known since the 1950s that nontoxigenic strains of Corynebacterium diphtheriae could become toxigenic after infection by temperate bacteriophages and a one-to-one correlation exists between the presence of the tox gene mechanism on a bacteriophage and subsequent DT production. The biology, structure, and molecular epidemiology of DT and the tox gene have been comprehensively reviewed (Holmes, 2000; Yates et al., 2006). Fragment A of the tox gene contains ADP-ribosyl-transferase activity; fragment B contains the receptor-binding and membrane-associating domains, with the structural tox gene being under the control of the chromosomal iron-dependant repressor, dtxR, and a promoter-operator region (Nakao et al., 1996).
Most nontoxigenic strains of Corynebacterium diphtheriae and most tox gene-negative bacteriophages do not contain detectable tox-related DNA sequences (Holmes, 2000). However, some strains contain detectable tox genes but do not express DT. It has been suggested that these strains have one or more of a variety dysfunctional genetic mechanisms which preclude DT production (von Graevenitz and Bernard, 2003); such isolates are considered as non-toxigenic with respect to public health response (Efstratiou et al., 1998; Efstratiou and George, 1999). The dtxR gene has been demonstrated to be heterogeneous, but all subtypes of this gene, if functional in strains lyosogenized by corynephage, could theoretically produce DT, and the possibility exists that non-toxigenic strains could revert to toxigenic status (De Zoysa et al., 2005).
Corynephage insert into the Corynebacterium diphtheriae chromosome at either one of two specific attachment sites, attB1 and attB2 (Oram et al., 2007; Seto et al., 2008). Protein transcribed by the DT repressor gene (dtxR), serves as a global repressor of metabolism in Corynebacterium diphtheriae. Cellular functions which are negatively regulated by iron (Fe2+) levels include production of DT, synthesis of a siderophore (also called corynebactin), corynebactin-dependant iron uptake, and utilization of iron from heme (Qian et al., 2002). DT catalyzes the NAD-dependant ADP ribosylation of elongation factor 2 and inhibits protein synthesis and death in cells from humans or susceptible animals (Qian et al., 2002).
Diphtheria-like disease in humans as manifested by Corynebacterium ulcerans, although long understudied, is now being described as an emerging, possibly zoonotic disease (Bonmarin et al., 2009; Hogg et al., 2009). Different opinions exist on the efficacy of diphtheria toxoid vaccine to protect against the effects of infection by toxigenic Corynebacterium ulcerans (Schuhegger et al., 2008a, 2008b). Differences (up to ∼5%) exist between nucleotide and amino acid sequences of Corynebacterium diphtheriae and Corynebacterium ulcerans tox genes (Sing et al., 2003); therefore, further study is required to clarify the efficacy of diphtheria toxoid to protect against disease caused by Corynebacterium ulcerans. Features of the attachment site in Corynebacterium ulcerans have been reviewed (Seto et al., 2008).
Complete genome analysis of a strain of Corynebacterium jeikeium, an opportunistic pathogen which is usually MDR and always lipophilic, revealed a 14,323-bp bacteriocin-producing plasmid which is designated pKW4. This analysis revealed that “lipophila” in Corynebacterium jeikeium, but probably in all other lipophilic species described in this section, is expression of a fatty acid auxotrophy due to the absence of fatty acid synthase (Tauch et al., 2005). This phenomenon was also observed from complete genome analysis of the lipophile Corynebacterium kroppenstedtii which, as a species which lacks mycolates, was also found to lack a mycolate reductase gene (Tauch et al., 2008a).
The role of plasmids, as bearers of virulence factors or in association with increased pathogenicity among Corynebacterium species, is poorly understood. The complete genome of an unusual black-pigmented strain of Corynebacterium aurimucosum (ATCC 700975, designated the type strain of Corynebacterium nigricans, CN-1T) that was associated with the female genital tract of a woman who experienced a spontaneous abortion during month 6 of her pregnancy, was described. A 29,037 bp plasmid designated pET44827 was found to code for a non-ribosomal peptide synthetase, which appeared to play a key role in the synthesis of the black pigment; this pigment was conjectured to play a role in both protecting the bacterium from the high hydrogen peroxide concentration of the vagina and play a role in causing complications in pregnancy (Trost et al., 2010).
Phospholipase D (Pld) is a major virulence factor of Corynebacterium pseudotuberculosis, the organism causing caseous lymphadenitis in sheep and goats in addition to being able to harbor corynephage and express DT. Pld is detected in Corynebacterium ulcerans as well as Arcanobacterium hemolyticum; in clinical microbiology laboratories, the presence of Pld is demonstrated by the inhibition of the Christie–Atkins–Munch–Petersen (CAMP) reaction (also called the reverse CAMP test) (Barksdale et al., 1981). Pld regulation and expression has been found to be a complex process which includes assisting with dissemination of the bacterium from site of infection to the lymph nodes and thereafter reducing macrophage viability (McKean et al., 2007). Rapid detection of Corynebacterium pseudotuberculosis among diseased animals has been made possible by the use of a multiplex assay to detect the 16S rRNA, rpoB, and pld genes of this bacterium (Pacheco et al., 2007).
Pathogenicity, virulence factors and ecological niches for other species remain understudied and largely unknown.
Antibiotic susceptibility and mechanisms of resistance
Antibiotic susceptibility of Corynebacterium species is less well studied compared with many other Gram-stain-positive rods because, for many years, “diphtheroids” were dismissed as contaminants and did not merit either the resources or interest required for further study. In addition, there was no consensus on standardized protocols and breakpoint interpretation for many years, so a wide variety of methods were independently used for clinically relevant isolates. Application of the broth microdilution method and specific MIC breakpoints were formally recommended by the CLSI in 2006 (Clinical Laboratory Standards Institute, 2006). However, Etest (e.g. Martinez-Martinez et al., 1995a), variations of agar dilution (e.g. Lagrou et al., 1998; Soriano et al., 1995), and disk diffusion (e.g. Riegel et al., 1996a; Weiss, et al., 1996) have also been used. A lack of precise species assignments and a lack of published historical data have made it difficult to compare older data to contemporary data to see if significant changes in resistance for specific taxa have occurred over time.
In 1971, Corynebacterium diphtheriae strains were found to be susceptible to commonly used drug classes tested, particularly those used for acute care treatment (penicillin and erythromycin) (McLaughlin et al., 1971). In 1995, diphtheria strains were found to be susceptible to beta lactams, third generation cephalosporins, vancomycin, pefloxacin, imipenam but not pefixime, azreonam, cefpodoxime, and some earlier generation cephalosporins; occasional strains were resistant to lincomycin and erythromycin (Patey et al., 1995). Antibiotic susceptibility patterns for species other than Corynebacterium diphtheriae confirmed observations that some or most strains of specific species, including Corynebacterium amycolatum, Corynebacterium striatum, Corynebacterium minutissimum, Corynebacterium afermentans, Corynebacterium argentoratense, Corynebacterium auris, Corynebacterium glucuronolyticum, lipophilic species Corynebacterium jeikeium, Corynebacterium tuberculostearicum (or as provisionally identified as CDC group G2), and Corynebacterium urealyticum, were often resistant to two or more drug classes (Funke et al., 1996b; Lagrou et al., 1998; Riegel et al., 1996a; Soriano et al., 1995; Williams et al., 1993). The new glycylcycline, tigecycline, and other commonly used antibiotics were evaluated against various Corynebacterium species, where tigecycline, as well as linezolid, daptomycin, vancomycin, and quinupristin/dalfopristin demonstrated good in vitro activity against isolates, but Corynebacterium coyleae and Corynebacterium aurimucosum strains were found to be MDR (Fernandez-Roblas et al., 2009). A daptomycin-resistant strain of Corynebacterium jeikeium has been reported (Schoen et al., 2009). Corynebacterium resistens was described as being susceptible only to vancomycin and minocycline but resistant to other drugs tested (Otsuka et al., 2005a).
In 1980, the presence of a 9.5 MDa plasmid was described as being associated with erythromycin resistance for strains of Corynebacterium diphtheriae (Schiller et al., 1980). In 1990, PCR was used to detect erm genes, i.e. genes encoding erythromycin ribosome methylases (rRNA methylases) that are linked to erythromycin resistance, from a variety of species (Arthur et al., 1990). Subsequently, studies of resistance mechanisms for various taxa have been reviewed including, among others, mef genes (macrolide efflux pump) and erm family genes, of which ermB, ermC, and particularly ermX have been detected among Corynebacterium species (Roberts, 2008). The ermX gene, believed to be borne on plasmids or transposons, adds one or two methyl groups to a single adenine in the 23S rRNA moiety and confers high level resistance to macrolides, lincosamides, and streptogramin B (MLS phenotype), as evidenced by resistance to at least erythromycin and clindamycin but often to other drug classes (Roberts, 2008). The ermX gene from Corynebacterium jeikeium has been linked to transposon Tn5432 for some, but not all, isolates studied (Rosato et al., 2001). To date, resistance as linked to the presence of ermX has been reported in Corynebacterium amycolatum and Corynebacterium jeikeium (Rosato et al., 2001; Tauch et al., 2005; Yague Guirao et al., 2005), Corynebacterium coyleae (Fernandez-Natal et al., 2008), Corynebacterium striatum (Campanile et al., 2009; Otsuka et al., 2006), Corynebacterium diphtheriae (Tauch et al., 2003), and Corynebacterium urealyticum (Tauch et al., 2008b). The ermX gene was not detected among complete genome studies of Corynebacterium kroppenstedtii and a black pigmented Corynebacterium aurimucosum strain (Tauch et al., 2008b; Trost et al., 2010). Resistance to quinolones, due to a mutation in the gyrA gene, was found among clinical isolates of Corynebacterium amycolatum and Corynebacterium striatum (Sierra et al., 2005). A single amino acid substitution at position 83 of the gyrA gene generated a norfloxacin-resistance phenotype, but double mutation with amino acid substitutions at positions 83 and 87 gave rise to high level resistance to other fluoroquinolones in a study of Corynebacterium macginleyi isolates involved in ocular infections (Eguchi et al., 2008). Corynebacterium striatum strains were found to possess ermX, tet A/B (related resistance, tetracycline, oxytetracycline, and oxacillin), cmx A/B (related resistance chloramphenicol) and aphA1 (related resistance, aminoglycoside) genes (Campanile et al., 2009). With time, other antibiotic resistance mechanisms among Corynebacterium species will be elucidated.
Ecology and habitat
There are at time of writing, ∼51 medically relevant species have been described that cause occasional infections in humans or are transmitted to humans by zoonotic means. Most are deemed to be rare opportunistic pathogens. Some species thought to be part of the common skin flora, such as Corynebacterium amycolatum, Corynebacterium jeikeium, and some of the other lipophilic species, have been found to be resistant to multiple drug classes and can cause significant and occasionally fatal disease, particularly in immunocompromised patients or nosocomially in hospitals or nursing homes (Funke and Bernard, 2007). However, many of the medically relevant species can also be recovered as commensals or contaminants from a variety of clinical specimens. Therefore, it is recommended that identification to species level should be attempted if the organism is isolated (i) from normally sterile body sites, e.g. blood (except if only one of multiple specimens became positive), (ii) from adequately collected clinical material if the Corynebacterium species is the predominant organism, and (iii) if recovered from urine specimens, e.g. Corynebacterium urealyticum (described further below), is the sole bacterium encountered with a bacterial count >104/ml or if it is the predominant organism recovered and the total bacterial count is >105/ml (Funke and Bernard, 2007).
Corynebacterium species can cause significant infection or appear to be commensals in animals or birds. Virulence factors and mechanisms of pathogenicity remain understudied. To date, 31 species associated with animals or birds have been described including: Corynebacterium amycolatum (mastitis in cattle) (Hommez et al., 1999), Corynebacterium aquilae and Corynebacterium falsenii (from eagles) (Fernández-Garayzábal et al., 2003), Corynebacterium auriscanis (otitis and suppurations in dogs) (Collins et al., 1999b), Corynebacterium bovis (mastitis and abscesses in cattle) (Watts et al., 2000), Corynebacterium camporealensis (mastitis in sheep) (Fernández-Garayzábal et al., 1998), Corynebacterium canis (dog mouth) (Funke et al., 2010a), Corynebacterium capitovis (sheep skin scrapings) (Collins et al., 2001a), Corynebacterium caspium (from a seal) (Collins et al., 2004), Corynebacterium ciconiae (stork trachea) (Fernández-Garayzábal et al., 2004), Corynebacterium cystitidis (pyelonephritis in cattle) (Yanagawa and Honda, 1978), Corynebacterium diphtheriae (mastitis, dermatitis, and wound infection in cattle or horses) (Corboz et al., 1996; Greathead and Bisschop, 1963; Henricson et al., 2000), Corynebacterium felinum (Scottish wild cat) (Collins et al., 2001b), Corynebacterium freiburgense (dog mouth) (Funke et al., 2009), Corynebacterium glucuronlyticum (genital tract of pigs) (Devriese et al., 2000), Corynebacterium kutscheri (rats, mice, and hamsters) (Collins and Cummins, 1986), Corynebacterium mastitidis (mastitis in sheep) (Fernández-Garayzábal et al., 1997). Corynebacterium minutissimum (mastitis in cattle) (Hommez et al., 1999), Corynebacterium mustelae (sepsis in ferrets) (Funke et al., 2010b), Corynebacterium phocae (nasal cavity of seal) (Pascual et al., 1998), Corynebacterium pilosum (urogenital tract of cattle) (Yanagawa and Honda, 1978), Corynebacterium pseudotuberculosis (ovine caseous lymphadenitis and other sheep or goat diseases) (Baird and Fontaine, 2007; Dorella et al., 2006), Corynebacterium renale (pyelonephritis in cattle) (Collins and Cummins, 1986), Corynebacterium sphenisci (from wild penguins) (Goyache et al., 2003a), Corynebacterium spheniscorum (from cloacae of wild penguins) (Goyache et al., 2003a), Corynebacterium suicordis (pericarditis, pneumonia, and lymph enlargements of pigs) (Vela et al., 2003), Corynebacterium testudinoris (from a tortoise) (Collins et al., 2001b), Corynebacterium ulcerans (goats, pigs, cattle, horses, cats, dogs, otters, and other) (Bonmarin et al., 2009; Schuhegger et al., 2009), Corynebacterium ulceribovis (bovine ulcers) (Yassin, 2009), Corynebacterium urealyticum (UTIs of dogs and cats) (Bailiff et al., 2005; Cavana et al., 2008), and Corynebacterium vitaeruminis (derived from cow rumen) (Collins and Cummins, 1986).
Corynebacterium species which have been recovered or detected in animals have also been documented to be potentially the cause of infection in humans by zoonotic transmission. These infections have been found to have occurred by occupationally related handling of the animals, by animal bites, or by unknown means. This has been observed with Corynebacterium auriscanis (Bygott et al., 2008), Corynebacterium bovis (Achermann et al., 2009; Bernard et al., 2002), Corynebacterium canis (Funke et al., 2009), Corynebacterium diphtheriae (Bonmarin et al., 2009), Corynebacterium freiburgense (Funke et al., 2009), Corynebacterium kutscheri (Holmes and Korman, 2007), a Corynebacterium mastitidis-like organism (Eguchi et al., 2008), Corynebacterium pseudotuberculosis (Bonmarin et al., 2009; Join-Lambert et al., 2006; Peel et al., 1997), and particularly of interest as an emerging zoonotic agent, toxigenic strains of Corynebacterium ulcerans (Bonmarin et al., 2009; De Zoysa et al., 2005b; Hogg et al., 2009; Wagner et al., 2001). Corynebacterium glucuronolyticum, Corynebacterium minutissimum, and Corynebacterium urealyticum (described further below) are usually associated with human disease, rather than animal disease, and evidence of transmission between human and animals has not been described.
Other Corynebacterium species have been described as being recovered from foodstuffs, the environment, or water, and some of these in turn have been widely used in industrial applications. These are Corynebacterium callunae (environment), Corynebacterium glutamicum (environment, production of glutamic acid), Corynebacterium flavescens (dairy products) (Collins and Cummins, 1986), Corynebacterium casei (soft cheeses) (Brennan et al., 2001), Corynebacterium doosanense (activated sludge) (Lee et al., 2009), Corynebacterium efficiens (environment, production of glutamic acid) (Fudou et al., 2002), Corynebacterium glaucum (cosmetic dye) (Yassin et al., 2003), Corynebacterium halotolerans (saline soil) (Chen et al., 2004), Corynebacterium lubricantis (coolant lubricant) (Kampfer et al., 2009), Corynebacterium maris (mucus of sea coral) (Ben-Dov et al., 2009), Corynebacterium marinum (sea sediment) (Du et al., 2010), Corynebacterium terpenotabidum (environment, capable of degrading squalene) (Takeuchi et al., 1999) and Corynebacterium variabile (formerly Arthrobacter variabilis, later Corynebacterium variabilis, from animal fodder) (Collins, 1987a).
Enrichment and isolation procedures
Members of this genus require one or more vitamins, amino acids, purines, and pyrimidines in culture medium to grow (Collins and Cummins, 1986; Funke and Bernard, 2007). Growth for some species is enhanced by the addition of lipids or by using an extended incubation period (detailed below, Corynebacterium urealyticum when recovered from urine). Growth is generally achieved using a temperature range from 30–37°C, particularly under a 5% CO2 atmosphere with other ranges described as appropriate by species, below. No species described to date will grow on MacConkey agar except Corynebacterium lubricantis (described below). Isolation of Corynebacterium diphtheriae, Corynebacterium ulcerans, or Corynebacterium pseudotuberculosis from non-sterile sites is outlined below.
Maintenance procedures
Corynebacterium species in general do not require special procedures for storage. Short-term (weeks or months) storage has historically been done using enriched media such as PAIs, Loeffler's, or blood agar slants (von Graevenitz and Bernard, 2003). Medium-term storage may be done by storage of bacteria on beads in cryopreservative at −80°C (available commercially), and long-term preservation may be done by standard lyophilization procedures.
Procedures for testing special characters
Contemporary biochemical procedures for testing medically relevant species of this genus have been outlined previously (Funke and Bernard, 2007; von Graevenitz and Bernard, 2003). Rapid identification panels such as the API Coryne strip (BioMérieux) have been described for characterization of many of these species and are especially useful for those which grow well within the allotted 48 h incubation period and are reactive with the slate of substrates. Users are cautioned that the underlying database is only infrequently updated and does not provide enough discrimination to delineate among newly described species outlined here. The API Coryne strip does not include starch utilization, a feature which historically was used in part to differentiate among biovars (= biotypes) of Corynebacterium diphtheriae. A web-based query to decode API data exists; it provides possible adjunct tests to assist with identification (https://apiweb.bioMerieux.com). Ancillary strips such as BioMérieux's API ZYM enzyme strip, API CH50 with API 20 E and panels manufactured by other companies, are also frequently used. The automated Biolog system has been used to provide phenotypic characteristics for various Corynebacterium species including the screening of nutritional and other physiological properties (Fernandez-Natal et al., 2009). Enzyme testing has been done as single test assays, such as described for pyrazinamidase or phosphatase (synonym of alkaline phosphatase) in a previous version of this chapter (Collins and Cummins, 1986). The characterization of species with no medical relevance that were derived from the environment has been carried out, either manually or by automated means (e.g. by use of the Biolog system) by testing the ability of the organism to utilize different carbon compounds, by defining optimal or preferred salinity, temperature, atmosphere, and pH, and by considering chemotaxonomic and genetic features; this has been outlined recently for Corynebacterium lubricantis (below) (Kampfer et al., 2009).
Metachromatic granules (synonym polymetaphosphate), the presence of which is described for some species in this text, are observed as bluish-purple after staining with methylene blue stain (Macfaddin, 2000).
Many species in this genus are lipophilic, i.e. they grow poorly at 35–37°C in 24 h or longer on standard laboratory media, but show enhanced growth in 48–72 h on sheep blood or brain heart infusion broth enriched with a lipid, such as 0.1–1.0% Tween 80, as detailed below for Corynebacterium macginleyi (Riegel et al., 1995e).
Special selective agars exist for Corynebacterium species, but the most widely used are those which select for Corynebacterium diphtheriae or Corynebacterium ulcerans from respiratory specimens, e.g. Tellerite or modified Tinsdale agars (Efstratiou and George, 1999).
The entire gene or fragments of the gene associated with the production of diphtheria toxin by Corynebacterium diphtheriae, Corynebacterium ulcerans, and Corynebacterium pseudotuberculosis can be detected by conventional (Efstratiou et al., 1998; Efstratiou et al., 2000) or real-time PCR (Mothershed et al., 2002; Schuhegger et al., 2008a, 2008b; Soriano et al., 2009). However, definitive toxigenic status is achieved only by detecting expression of diphtheria toxin, which is generally done using the modified Elek test (Engler et al., 1997). CAMP reaction and expression of phospholipase D by the CAMP inhibition assay for Corynebacterium ulcerans and Corynebacterium pseudotuberculosis strains (Barksdale et al., 1981) are useful tests for these species.
Differentiation of the genus Corynebacterium from other genera
Of the 60-plus Corynebacterium species described since publication of the previous edition of the Manual, many but not all authors describe testing for the presence of corynemycolates, types of cell-wall sugars and/or types of diaminopimelic acid, types and quantities of long-chain cellular fatty acids, and G+C mol% (Table 1). Testing for other features such as for other types of cell-wall lipids or phospholipids, polyamines, or acyl types are now only infrequently done. Staining for the presence of metachromatic granules is now almost never described.
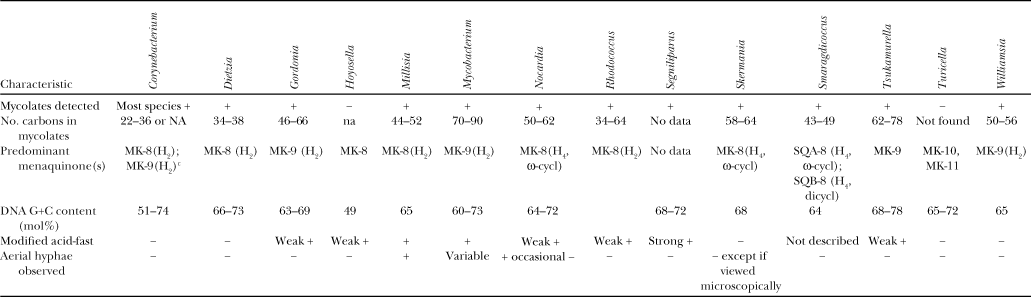
- a Symbols: +, >85% positive; −, 0–15% positive; na, not applicable; some small discrepancies exist among references describing no. of carbons in mycolates, so broadest range used (data from Chun et al., 1997; Kämpfer et al., 1999; Soddell et al., 2006; Adachi et al., 2007; Jurado et al., 2009; Koerner et al., 2009).
- b Genera of the order Corynebacteriales; all genera have cell-wall type IV, that is with meso-diaminopimelic acid, arabinose, and galactose with the caveat that CW sugars were not determined for the genus Segniliparus (Butler et al., 2005); Corynebacterium, Dietzia, and Hoyosella have an acetylated acyl type; the remaining genera have glycolated acyl types except for Segniliparvus and Turicella, where acyl type has not been determined.
- c MK-7 (H2) also detected in Corynebacterium lubricantis (Kampfer et al., 2009) and MK-10(H2) found in Corynebacterium thomssenii (Zimmermann et al., 1998).
Taxonomic comments
The genus Corynebacterium belongs to the phylum Actinobacteria, class Actinobacteria, order Corynebacteriales, family Corynebacteriaceae. Specific 16S rRNA signature nucleotides for these bacteria have been outlined (Zhi et al., 2009).
It has become increasingly difficult to distinguish Corynebacterium species based on phenotypic testing alone, as shown from some of the data outlined in Table 2. Additional identification approaches, such as the use of 16S rRNA and rpoB gene sequencing, are being used more routinely as a significant adjunct to characterization. However, assignment of species nova to this genus requires testing for as many genetic and chemotaxonomic properties as are available to researchers, and, increasingly, full genome sequencing is highly recommended.
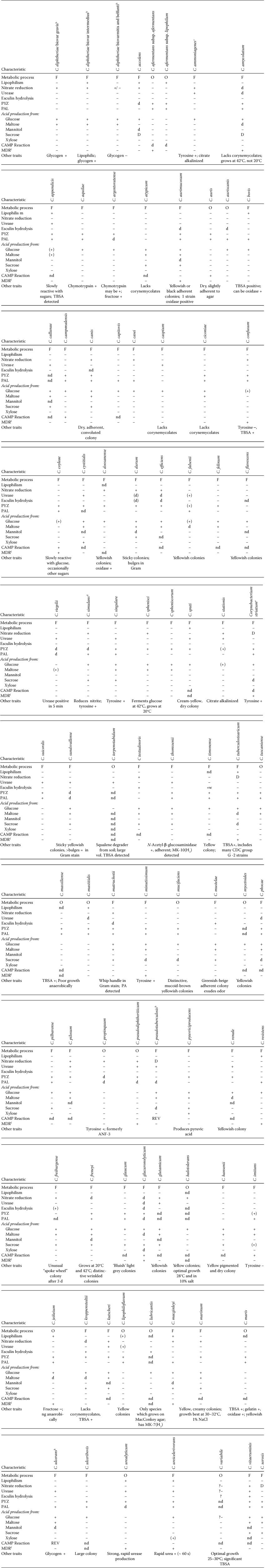
- a Species recovered from human clinical material except as described in comment. Abbreviations and symbols: F, fermentative; O, oxidative; +, positive; −, negative; d, variable; ( ), delayed or weak reaction; nd, no data for test described; ng, no growth; REV, CAMP inhibition reaction; PYZ, pyrazinamidase; PAL, alkaline phosphatase and in older texts, was also called phosphatase; unk, unknown; TBSA, cellular fatty acid tuberculosteric acid (10-methyl 18:0) detected.
- b Corynebacterium diphtheriae biovar mitis reduces nitrate but biovar belfanti is nitrate negative; all biovars of Corynebacterium diphtheriae, Corynebacterium pseudotuberculosis, and Corynebacterium ulcerans produce cysteinase which is detected by producing a brown-black colony with a brown black halo on cysteine-containing agar such as Tinsdale medium. These species can be lyogenized by the bacteriophage that confers ability to produce diphtheria toxin.
- c Corynebacterium ammoniagenes (as Brevibacterium ammoniagenes) described as being completely non-reactive in sugars, and that reactivity was media dependant (Jones and Keddie, 1986).
- d Corynebacterium simulans, a strong nitrite reducer at low and high concentrations may appear to be nitrate reduction negative unless further tested using zinc dust (Wattiau et al., 2000). One strain of this species was catalase negative (Bernard et al., 2002).
- e For Corynebacterium striatum, the CAMP reaction, if done, has been described as negative (Martinez-Martinez et al., 1995b) or occasionally positive (Leonard et al., 1994).
- f MDR, Multidrug resistance described at least once in the literature as being resistant to three or more drug classes using CLSI guidelines and current data (Clinical Laboratory Standards Institute, 2006, 2009).
Many of the genera described in the previous edition of this Manual as being closely related to the genus Corynebacterium have subsequently been found to be unrelated or qualitatively distinguishable by genetic and chemotaxonomic methods. All Corynebacterium species previously attributed to causing disease in plants have been reassigned, and none of the species described here have been documented to be phytopathogens. Corynebacterium species reassigned since the previous edition to other genera include: Corynebacterium betae, Corynebacterium oortii, Corynebacterium flaccumfaciens Hedges 1922 and Corynebacterium poinsettiae to Curtobacterum flaccumfaciens comb. nov. Collins and Jones 1984 (Effective publication: Collins and Jones 1983b.); Corynebacterium equi and Corynebacterium hoagii to Rhodococcus equi (Magnusson 1928) Goodfellow and Alderson 1977AL; Corynebacterium fascians Tilford 1936 to Rhodococcus fascians comb. nov. Goodfellow 1984b (Effective publication: Goodfellow 1984a.); Corynebacterium insidiosum, Corynebacterium michiganense, Corynebacterium nebraskense, and Corynebacterium sepedonicum to the genus Clavibacter as subspecies of Clavibacter michiganensis (Davis et al. 1984); Corynebacterium iranicum to Rathayibacter iranicus comb. nov. Zgurskaya et al. 1993; Corynebacterium paurometabolum Steinhaus 1941 to Tsukamurella paurometabolum comb. nov. Collins et al. 1988c, later corrected to Tsukamurella paurometabola by Euzéby (1998); Corynebacterium pyogenes Glage 1903 to Actinomyces pyogenes Reddy et al. 1982 to Arcanobacterium pyogenes comb. nov. (Glage 1903) Pascual Ramos et al. 1997 and then to Trueperella pyogenes comb. nov. (Glage 1903) Yassin et al. 2011; Corynebacterium rathayi Smith 1913 to Clavibacter rathayi comb. nov. Davis et al. 1984 then to Rathayibacter rathayi comb. nov. (Smith 1913) Zgurskaya et al. 1993, and lastly, Corynebacterium tritici Carlson and Vidaver 1982 to Clavibacter tritici comb. nov. Davis et al. 1984 then to Rathayibacter tritici comb. nov. (Carlson and Vidaver 1982) Zgurskaya et al. 1993.
Corynebacterium ilicis was proposed as the name for a bacterial pathogen of American holly that caused a blight of foliage and twigs (Mandel et al., 1961). The type strain (DSM 20138 = ATCC 14264 = NCPPB 1228) was belatedly designated in 1977 (Dye and Kemp, 1977). Assignment of Corynebacterium ilicis to the genus Corynebacterium was controversial and subsequently it was reclassified as Arthrobacter ilicis Collins et al. 1981. Upon further review, the Corynebacterium ilicis type strain was found to be apathogenic for plants and a member of a different taxon group with respect to Corynebacterium ilicis reference strains ICMP 2608 and ICMP 2609, described at the same time as the type strain, which were pathogenic for American holly. Following Request for an Opinion no. 87 (Young et al., 2004), the Judicial Commission ruled that features associated with the name Corynebacterium ilicis Mandel et al. 1961 should be represented by the type strain ICMP 2608 = ICPB CI144 (later reassigned to Curtobacterium flaccumfaciens), and that Arthrobacter ilicis would be typified by strain DSM 20138 = ATCC 14264 = NCPPB 1228 (Effective publication: Collins et al., 1981.) and validated (Collins et al., 1982c; Young et al., 2004; Judicial Commission of the International Committee on the Systematics of Prokaryotes, 2008).
Brevibacterium liquefaciens Okabayashi and Masuo 1960AL was reclassified as Corynebacterium liquefaciens (Okabayashi and Masuo 1960) Lanéelle et al. 1980 based on the study of the type strain, ATCC 14929 (Lanéelle et al., 1980). This isolate was later found to have significant differences when compared to a new copy of ATCC 14929 and, as a result, all recommendations for reassignment were withdrawn in an erratum. Brevibacterium liquefaciens strain ATCC 14929 was subsequently reassigned to Arthrobacter nicotianae Giovannozzi-Sermanni 1959 by Gelsomino et al. (2004).
It is not anticipated that further changes in taxonomic classification of members of the genus Corynebacterium will occur except by the emendation or augmentation by new species.
List of species of the genus Corynebacterium
Information on this list was corroborated with by information compiled by Euzéby (2010). Accession numbers for rpoB gene sequences describe either complete gene (3100–3450 bp) or partial gene sequences (with ∼400–425 bp).
Corynebacterium diphtheriae
(Kruse 1886) Lehmann and Neumann 1896, 350AL (“Bacillus diphtheria” Kruse in Flügge 1886, 225)
diph.the.ri'a.e. Gr. n. diphthera leather, skin; N.L. fem. n. diphtheria a disease in which a leathery membrane forms in the throat; N.L. gen. n. diphtheriae of diphtheria.
(This description is largely based on that of Collins and Cummins, 1986.)
Straight or slightly curved rods, frequently swollen at one or both ends, 0.3–0.8 × 1.0–8.0 µm. Usually stains unevenly and often contains metachromatic granules (polymetaphosphate) which stain bluish purple with methylene blue. Gram-stain-positive, but easily decolorized, especially in older cultures.
Descriptive cultural types or biovars (= biotypes) of Corynebacterium diphtheriae strains are still commonly applied and are designated gravis, intermedius, mitis, and belfanti. Designations for gravis, intermedius, and mitis were originally given in accordance with the clinical severity of cases from which the different strains were most frequently isolated. On blood agar, colonies vary in size and appearance but generally are 1–3 mm at 24 h (except intermedius, described below) and may demonstrate a narrow band of hemolysis, but no soluble hemolysin is produced. Colonies on blood tellurite (with 0.04% potassium tellurite) are gray to black, with appearance dependant on type. Colonies on modified Tinsdale agar are 1–2 mm, black or charcoal gray, with each colony having a brown-black halo visible in the agar as cysteinase produced by the organism reacts with the cysteine in the medium. Although less frequently used now, growth on Loeffler's medium is abundant, with grayish to cream colored colonies and no liquefaction. The use of historical descriptions of colonies (such as gravis being large, radially striated brittle colonies and mitis being smooth, shiny, and butyrous) or Gram stain details (gravis having short regular rods, intermedius with long rods with marked cross-striations, mitis with long curved irregular rods) as a means to subdivide strains into gravis, intermedius, and mitis are now rarely used.
Biovars are usually reported for clinical isolates and are generally differentiated by phenotype: biovar gravis reduces nitrate and utilizes both glycogen and starch; biovar intermedius is somewhat lipophilic in that colonies are 0.5–1 mm after 24–48 h growth on blood agar, but otherwise reduces nitrate and may utilize glycogen (rapid strip API Coryne usually negative but conventional tube sugar positive) and starch; biovar mitis reduces nitrate but does not utilize glycogen and rarely starch; biovar belfanti neither reduces nitrate nor utilizes glycogen or starch. CAMP and CAMP inhibition reactions negative. All strains exhibit the same reactions except for occasional sucrose-fermenting isolates described primarily in South America (de Mattos-Guaraldi and Formiga, 1998) or Corynebacterium diphtheriae biovar belfanti like isolates that were derived from symptomatic cats and did not ferment maltose (Hall et al., 2010). Optimal growth temperature 30–37°C, and complex media is required for growth.
All Corynebacterium diphtheriae strains, regardless of biovar, conform to the genus description chemotaxonomically. All contain corynemycolates, the polar lipids phosphatidylglycerol, phosphatidylinositol, and monoacylated phosphatidylinositol dimannoside, and MK-8 (H2) as major menaquinone. Cell wall contains meso-diaminopimelic acid and the sugars arabinose and galactose. Straight-chain saturated fatty acids are mainly palmitic and stearic acids with minimal branched saturated fatty acids; significant quantities of C16:1 ω7c isomer is characteristically detected (Bernard et al., 1991). All produce propionic as a product of fermentation (Bernard et al., 2002).
Source: clinical specimens from humans or animals.
DNA G+C content (mol%): 52–55 (method unknown); 53.48 (full genome sequencing).
Type strain: ATCC 27010, CIP 100721, DSM 44123, NCTC 11397.
Sequence accession no. (16S rRNA gene): GQ118341, X82059, X84248.
Further comments: additional GenBank accession numbers include (complete genome sequence) BX248353 and (complete rpoB gene) AY492230. All biovars of Corynebacterium diphtheriae may produce diphtheria toxin which clinically should be tested for as described elsewhere in this chapter. Non-toxigenic strains otherwise have typical features. Full genome sequence of Corynebacterium diphtheriae strain NCTC 13129 has been published containing 2,488,635 bp (Cerdeno-Tarraga et al., 2003).
Corynebacterium accolens
Neubauer, Sourek, Ryc, Bohacek, Mara and Mnukova 1991a, 331VP (Effective publication: Neubauer, Sourek, Ryc, Bohacek, Mara and Mnukova 1991b, 50.)
ac'co.lens. L. v. accolere live by; L. part. adj. accolens living close by.
Cells are Gram-stain-positive rods, 0.6–0.9 × 1.0–2.0 µm, nonmotile, occurring singly or in clusters typical for corynebacteria. Colonies are roundish, slightly convex, grayish, and show the phenomenon of satellitism, i.e., enhanced growth in the vicinity of Staphylococcus aureus indicating that the strains are lipophilic. Facultatively anaerobic. Oxidase negative. Growth occurs at 30–40°C with optimal growth at 37°C, pH 7.2. Acid is produced from D-glucose, galactose, fructose, D-mannose, and maltose. Fermentation of sucrose is variable. Acid is not produced from lactose, D-xylose, trehalose, L-arabinose, raffinose, L-rhamnose, dextrin, mannitol, dulcitol, D-sorbitol, glycerol, myo-inositol, salicin, and inulin. Esculin, urea, gelatin, casein, and starch are not hydrolyzed. No production of indol, H2S, acetoin, and lipase. The methyl red test is negative. CAMP reaction negative. Cell wall contains meso-diaminopimelic acid; corynemycolic acids made up of 22–36 carbon atoms and arabinose and galactose sugars. Cellular fatty acids are as described for the genus. Menaquinone or polar lipid types not extant. Does not produce propionic acid as metabolic product (Bernard et al., 2002).
Source: human clinical specimens, in particular specimens from ear, nose, throat, and eyes.
DNA G+C content (mol%): 53.2 (method unknown).
Type strain: CNCTC Th1/57, ATCC 49725, CCUG 28779, CIP 104783, DSM 44278, JCM 8331.
Sequence accession no. (16S rRNA gene): AJ439346, X80500.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492242. rpoB gene sequencing should be done to distinguish strains from Corynebacterium macginleyi, as those species can not be easily distinguished by 16S rRNA gene sequencing.
Corynebacterium afermentans
Riegel, de Briel, Prévost, Jehl, Monteil and Minck 1993b, 291VP
a.fer.men'tans. Gr. pref. a not; L. part. adj. fermentans leavening; N.L. part. adj. afermentans nonfermenting; i.e. nonfermenting carbohydrates.
The bacteria are Gram-stain-positive irregular rods or coccobacilli that sometimes contain metachromatic granules. Cells are arranged in typical V-shaped forms or palisades. Nonlipophilic. Good growth under aerobic conditions, and very slight growth under anaerobic conditions. Smooth, grayish-white colonies with entire margins. Colonies are 1–2 mm in diameter. Acid is not produced from glucose, glycogen, lactose, sucrose, ribose, D-xylose, D-mannose, D-galactose, trehalose, and D-mannitol. Cells utilize acetate and lactate. Propionate, D-glucose, L-arabinose, D-mannose, D-mannitol, N-acetylglucosamine, maltose, gluconate, caprate, adipate, malate, citrate, and phenylacetate are not utilized. Nitrate is not reduced. Urea and esculin not hydrolyzed. Tyrosine, gelatin, DNA, and starch are not degraded. The methyl red test is negative, and acetoin, indole, and H2S (on triple-sugar iron agar) are not produced. Hydrolysis of hippurate is variable. Growth is visible in 6.5% NaCl. Alkaline phosphatase, esterase, lipase, and acid phosphatase are produced. α-Galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, and β-glucosidase are not produced. All strains are resistant to fosfomycin. CAMP reaction variable (Funke and Bernard, 2007).
The cell wall contains meso-diaminopimelic acid, arabinose, and galactose. Cellular fatty acids are consistent with those described for the genus. Corynemycolates are present with C30:0, C30:1, C32:1, C33:1, C34:2, and C36:2 predominating. Does not produce propionic acid as metabolic product (Bernard et al., 2002).
Source: human clinical specimens.
DNA G+C content (mol%): 66 (HPLC).
Type strain: ATCC 51403, CCUG 32103, CIP 103499, DSM 44280, JCM 10390, LCDC 88199.
Sequence accession no. (16S rRNA gene): X82054.
Further comments: Corynebacterium afermentans Riegel et al. (1993b) was previously known as CDC coryneform ANF-1. By 16S rRNA gene sequencing, Corynebacterium afermentans is closely related to (98% identity with) to Corynebacterium coyleae and Corynebacterium mucifaciens but differs (∼94% identity with) when rpoB gene sequences are compared (Khamis et al., 2004) and by phenotypic testing (Table 2).
Corynebacterium afermentans subsp. afermentans
Riegel, de Briel, Prévost, Jehl, Monteil and Minck 1993b, 291VP
a.fer.men'tans. Gr. pref. a not; L. part. adj. fermentans leavening; N.L. part. adj. afermentans nonfermenting; i.e. nonfermenting carbohydrates.
Colonies are 1–2 mm in diameter after 24 h on blood agar and are not lipophiilc.
Source: human clinical specimens.
DNA G+C content (mol%): 66 (HPLC).
Type strain: ATCC 51403, CCUG 32103, CIP 103499, DSM 44280, JCM 10390, LCDC 88199.
Sequence accession no. (16S rRNA gene): X82054.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492265.
Corynebacterium afermentans subsp. lipophilum
Riegel, de Briel, Prévost, Jehl, Monteil and Minck 1993b, 291VP
li.po'phi.lum. Gr. n. lipos animal fat; Gr. neut. adj. philon loving; N.L. neut. adj. lipophilum fat loving.
The characteristics of Corynebacterium afermentans subsp. lipophilum are similar to those of Corynebacterium afermentans subsp. afermentans except that very small colonies (<0.5 mm in diameter) appear after 48 h on sheep blood agar. Strains are lipophilic. Hippurate is not hydrolyzed.
Chemotaxonomic features are like those of Corynebacterium afermentans subsp. afermentans except that branched saturated fatty acids are heptadecanoic acid, and straight-chain unsaturated fatty acids are mainly oleic acid, linoleic acid, and arachidonic acid.
Source: human clinical specimens.
DNA G+C content (mol%): 68 (HPLC).
Type strain: T18502, ATCC 51404, CCUG 32105, CIP 103500, DSM 44282, JCM 10391.
Sequence accession no. (16S rRNA gene): X82055.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492266.
Corynebacterium ammoniagenes
(Cooke and Keith 1927) Collins 1987b, 442VP (“Bacterium ammoniagenes” Cooke and Keith 1927)
am.mo.ni.a'ge.nes. N.L. n. ammonia ammonia; N.L. suff. -genes (from Gr. v. gennaô to produce) producing; N.L. part. adj. ammoniagenes ammonia-producing.
Gram-stain-positive, nonsporeforming, nonmotile rod-shaped cells (1–4.5 × 0.6–1.2 µm in diameter). Cells are irregular and occur singly or in pairs and may exhibit V forms. Colonies are circular, low convex with an entire edge, and are gray-white or yellow. Facultatively anaerobic. Optimum growth temperature is approximately 30°C. Growth occurs at 10°C and 37°C but not at 45°C. Growth occurs in the presence of 10% NaCl. Fermentation of carbohydrates has been described as being equivocal (Collins, 1987b). In a recent study, the following reactions were found for ATCC 6871T: glucose, ribose, and fructose were slowly fermented, but xylose, mannitol, lactose, maltose, galactose, glycerol, glycogen, raffionse, salicin, trehalose, and mannose were not. Nitrate reduced to nitrite. Urease is produced. Hippurate and tyrosine are hydrolyzed. Gelatin, casein, DNase, and starch are not hydrolyzed. Citrate is alkalinized. CAMP and reverse CAMP reactions are negative. Leucine arylamidase positive, but pyrazinamidase, pyrrolidonyl arylamidase, alkaline phosphatase, esterase, esterase lipase, lipase, valine arylamidase, cystine arylamidase, trypsin, alpha chymotrypsin, acid phosphatase, naphthol-AS-Bl-phosphohydrolase, α-galactosidase, β-galactosidase, β-glucuronidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase not detected (Bernard et al., 2010). Some organic acids may be assimilated.
Cell wall contains meso-DAP. The glycan moiety of murein contains only acetyl residues. Cell wall contains arabinogalactan polymer. Corynomycolic acids are present. Long-chain fatty acids are of the saturated, monounsaturated, and 10-methyl-branched types with hexadecanoic, octadecanoic, octadecenoic, and 10-methyl-octadecanoic acids predominating. TBSA detected but does not produce propionic acid as metabolic product (Bernard et al., 2010). The major phospholipids are diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol, and phosphatidylinositol dimannoside. The major menaquinone is MK-9(H2).
Source: feces of infants and piggery waste.
DNA G+C content (mol%): 53.7–55.8 (Tm).
Type strain: ATCC 6871, CCUG 38796, CIP 101283, DSM 20306, JCM 1305, NBRC 12612, NCCB 60030, NCIMB 8143, VKM B-672.
Sequence accession no. (16S rRNA gene): X84440.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492243 and (partial rpoB gene) FJ648511.
Corynebacterium amycolatum
Collins, Burton and Jones 1988b, 449VP (Effective publication: Collins, Burton and Jones 1988a, 351.)
a.my.co'la.tum. N.L. neut. adj. amycolatum wanting in mycolates.
Cells are Gram-stain-positive, nonmotile, nonsporeforming, pleomorphic rods. Colonies are whitish-grayish, dry, rough, and with uneven edges. Facultatively anaerobic. Nonlipophilic. Grows at 37°C but not at 10°C or 20°C at 3 d (Wauters et al., 1998); some strains grow at 40°C. Grows in 5% NaCl; some strains grow in 10% NaCl. Acid is usually produced from D-fructose, D-glucose, glycerol, D-mannose, and ribose. Acid production from maltose, sucrose, galactose (Wauters et al., 1996), and trehalose is variable. Acid is not produced from N-acetylglucosamine, starch, amygdalin, D-arabinose, L-arabinose, D-arabitol, L-arabitol, arbutin, cellobiose, D-fucose, L-fucose, b-gentiobiose, glycogen, gluconate, 2-keto-gluconate, 5-keto-gluconate, inositol, inulin, lactose, D-lyxose, mannitol, melezitose, melibiose, α-methyl-D-glucoside, α-methyl-D-mannoside, β-methyl xyloside, D-raffinose, rhamnose, salicin, sorbitol, L-sorbose, xylitol, D-xylose, and L-xylose. Glucose may be fermented at 42°C and ethylene glycol may be acidified at 48 h (Wauters et al., 1998). CAMP reaction is negative. Nitrate reduction and urea hydrolysis are variable. Esculin, arginine, casein, cellulose, gelatin, ornithine, tyrosine, and xanthine are not hydrolyzed. Starch is hydrolyzed (Funke et al., 1996a). Some strains hydrolyze hippurate. Pyrazinamidase, alkaline phosphatase, esterase, esterase lipase, cystine arylamidase, and acid phosphatase are detected; lipase, leucine arylamidase, and phosphoamidase are variable, but valine arylamidase, trypsin, chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase are not detected (Funke et al., 1996a). Voges–Proskauer and methyl red positive.
Cell-wall peptidoglycan contains meso-diamimopimelic acid, alanine, and glutamic acid. Arabinose and galactose are wall sugars. Mycolic acids are not present. The cellular fatty acids are consistent with those described for the genus. The principle menaquinone is MK-9(H2) with significant quantities of MK-8(H2). Polar lipid types include acyl phosphatidylglycerol, phosphatidylinositol, phosphatidylinositol dimannoside, and several unknown phospholipids (Collins et al., 1988a; Yague et al., 1997). Strains produce propionic acid as a product of fermentation (Funke et al., 1996a).
Source: human clinical specimens; also recovered from animals (Hommez et al., 1999).
DNA G+C content (mol%): 61 (Tm).
Type strain: S160, ATCC 49368, CCUG 35685, CIP 103452, DSM 6922, JCM 7447, NBRC 15207, NCIMB 13130.
Sequence accession no. (16S rRNA gene): X82057.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492241. When identification is done based solely on the use of phenotypic methods, Corynebacterium amycolatum isolates have been misidentified as Corynebacterium xerosis, Corynebacterium minutissimum, Corynebacterium striatum, and other closely related taxa (Funke et al., 1996a; Wauters et al., 1996; Zinkernagel et al., 1996). Most strains identified as Corynebacterium amycolatum using a polyphasic approach are MDR due in part to the presence of the ermX gene and other mechanisms as yet not fully elaborated (Yague et al., 1997). Older publications where identification of Corynebacterium xerosis and Corynebacterium minutissimum was based on biochemical, not genetic, traits should be interpreted with caution, as strains described as being multidrug resistant may, in fact, be misidentified Corynebacterium amycolatum isolates. The Corynebacterium amycolatum type strain (CCUG 35685T) and at least one other reference strain, CIP 100836 (“Corynebacterium asperum”), differ from most clinical isolates as they are fully sensitive to all drug classes tested by microbroth dilution (K. Bernard, personal communication). Strains formerly designated CDC group I2 or CDC group F2 have been assigned to Corynebacterium amycolatum (Wauters et al., 1996).
Corynebacterium appendicis
Yassin, Steiner and Ludwig 2002b, 1168VP
ap.pen'di.cis. L. fem. gen. n. appendicis of an appendage, intended to mean pertaining to appendicitis, from which the patient from whom the clinical material was taken for isolation of the organism was suffering.
Cells are Gram-stain-positive, thin, nonmotile, nonsporeforming, and pleomorphic coryneforms. Form small (<0.5 mm in diameter), grayish, and nonhemolytic colonies on Columbia sheep blood agar after 2 d at 37°C. Lipophilic. Facultatively anaerobic. Nitrate is not reduced. Acid is produced from glucose, maltose, and glycerol but not from arabinose, arabitol, cellobiose, glycogen, inulin, lactose, mannitol, melibiose, melzitose, pullulan, salicin, sorbitol, sucrose, tagatose, trehalose, raffinose, rhamnose, ribose, or xylose. Does not hydrolyze esculin, gelatin, hippurate, or starch. Displays urease, alkaline phosphatase, and pyrazinamidase activities but not acid phosphatase, arginine dihydrolase, esterase, esterase lipase, lipase, α-glucosidase, β-glucosidase, α-galactosidase, β-galactosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, α-mannosidase, α-fucosidase, phosphoamidase, alanine-phenylalanine-proline arylamidase, glycyl-tryptophan arylamidase, pyroglutamic acid arylamidase, pyrrolidonyl arylamidase, valine arylamidase, cystine arylamidase, leucine arylamidase, trypsin, and chymotrypsin. Acetoin production is positive, but indole production is negative. Produces lactate, but not propionate, as the major product of glucose fermentation.
Contains meso-DAP as wall diamino acid in addition to galactose and arabinose in whole-cell hydrolysates (i.e. cell-wall chemotype IV). Corynomycolic acids are present, and the fatty acid profile contains saturated, unsaturated, and huge amounts of tuberculostearic acids. Type PI phospholipids pattern with no nitrogen-containing compounds.
Source: abdominal swab of a patient with appendicitis with abscess formation.
DNA G+C content (mol%): 65.8 (HPLC).
Type strain: CCUG 48298, DSM 44531, IMMIB R-3491, JCM 11765, NRRL B-24151.
Sequence accession no. (16S rRNA gene): AJ314919.
Further comments: additional GenBank accession numbers include (partial rpoB gene) EU004066.
Corynebacterium aquilae
Fernández-Garayzábal, Egido, Vela, Briones, Collins, Mateos, Hutson, Domínguez and Goyache 2003, 1137VP
a.qui'la.e. L. gen. n. aquilae of an eagle.
Cells are Gram-stain-positive, nonmotile, nonsporeforming rods. Colonies are whitish, low convex, nonhemolytic, dry, and rough, 1–2 mm in diameter after 48 h incubation at 37°C. Facultatively anaerobic. Oxidase negative. Nonlipophilic. CAMP reaction is negative. Nitrate is not reduced. Acid is produced from D-glucose, D-fructose, D-mannose, glycerol, ribose, N-acetylglucosamine, and galactose, but not from maltose, trehalose, D-xylose, L-xylose, mannitol, lactose, sucrose, erythritol, D-arabinose, L-arabinose, adonitol, methyl β-xyloside, L-sorbose, rhamnose, inositol, sorbitol, methyl α-D-mannoside, methyl α-D-glucoside, amygdalin, arbutin, salicin, cellobiose, melibiose, inulin, melezitose, D-raffinose, xylitol, b-gentiobiose, D-turanose, D-lyxose, D-tagatose, D-fucose, L-fucose, D-arabitol, L-arabitol, 2-keto-gluconate, 5-keto-gluconate, and glycogen. Esculin, urea, and gelatin are not hydrolyzed. Alkaline and acid phosphatases, esterase, esterase lipase, leucine arylamidase, chymotrypsin, and phosphoamidase activities are detected. No activity is detected for lipase, α-glucosidase, β-glucosidase, β-glucuronidase, α-mannosidase, α-galactosidase, β-galactosidase, α-fucosidase, N-acetyl- β-glucosaminidase, valine arylamidase, cystine arylamidase, and trypsin.
The cell wall contains meso-DAP. Mycolic acids (C30-C36) are present. Fatty acids are of the straight-chain saturated (C14:0, C16:0, and C18:0) and monounsaturated (C16:1 ω9c and C18:1 ω9c) types.
Source: eagles.
DNA G+C content (mol%): not available.
Type strain: S-613, CCUG 46511, CECT 5993, JCM 12268.
Sequence accession no. (16S rRNA gene): AJ496733.
Corynebacterium argentoratense
Riegel, Ruimy, de Briel, Prévost, Jehl, Bimet, Christen and Monteil 1995a, 537VP
ar.gen.to.ra.ten'se. L. neut. adj. argentoratense of or pertaining to Argentoratus, Latin name of the city of Strasbourg, where the organism was isolated.
Cells are Gram-stain-positive, nonmotile, nonsporeforming pleomorphic rods. They grow on blood agar as circular acuminated creamy colonies which are not hemolytic. Facultatively anaerobic. Nonlipophilic. Nitrate is not reduced. Esculin, urea, gelatin, tyrosine, and DNA are not hydrolyzed or degraded. Hydrolyses of starch and hippurate occur variably. Acid is produced from D-glucose and fructose but not from sucrose, maltose, lactose, galactose, D-xylose, trehalose, glycogen, or D-mannitol. Acidification from ribose occurs variably. Pyrazinamidase, esterase lipase, cystine arylamidase, and, characteristically, α-chymotrypsin are present. Production of esterase, leucine arylamidase, and acid phosphatase is variable, and alkaline phosphatase is usually not produced. Pyrrolidonyl arylamidase, β-glucuronidase, β-galactosidase, α-glucosidase, and N-acetyl- β-glucosaminidase are not produced.
The cell wall contains meso-DAP and the sugars arabinose and galactose. Mycolic acids of short-chain lengths (C26–C36) are present. Strains produce propionic acid as a metabolic product (Bernard et al., 2002).
Source: human clinical specimens, in particular throat specimens.
DNA G+C content (mol%): 60–61 (HPLC).
Type strain: IBS B10697, ATCC 51927, CCUG 34893, CIP 104296, DSM 44202, JCM 10392.
Sequence accession no. (16S rRNA gene): X83955.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492249.
Corynebacterium atypicum
Hall, Collins, Hutson, Lawson, Falsen and Duerden 2003, 1067VP
a.ty'pi.cum. N.L. neut. adj. atypicum not typical, referring to the absence of corynomycolic acids, a feature normally present in corynebacteria.
Cells are Gram-stain-positive, short to filamentous rods. Colonies on Columbia agar with 5% horse blood, after incubation at 37°C for 48 h, are pinpoint, convex, entire-edged, shiny, white, and nonhemolytic. Facultatively anaerobic. Nonlipophilic. Growth occurs in broth containing 7.5% NaCl but not in 10% NaCl. Nitrate is not reduced to nitrite. Esculin, urea, gelatin, and starch are not hydrolyzed. Acid is produced from D-glucose, maltose, ribose, and sucrose but not from lactose, mannitol, glycogen, or D-xylose. Activity is detected for β-glucuronidase, cystine arylamidase, leucine arylamidase, and valine arylamidase. Alkaline and acid phosphatases, chymotrypsin, esterase, esterase lipase, α-fucosidase, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, lipase, α-mannosidase, N-acetyl-β-glucosaminidase, phosphoamidase, pyrrolidonyl arylamidase, pyrazinamidase, and trypsin are not produced. Acetoin is not produced.
The cell-wall murein contains meso-DAP. Cellular fatty acids are consistent with those described for the genus, but tuberculostearic acid is not present. Mycolic acids are not detected.
Source: an unknown human clinical source; habitat is not known.
DNA G+C content (mol%): not available.
Type strain: R2070, CCUG 45804, CIP 107431, JCM 12368.
Sequence accession no. (16S rRNA gene): AJ441057.
Corynebacterium aurimucosum
Yassin, Steiner and Ludwig 2002a, 1004VP emend. Daneshvar, Hollis, Weyant, Jordan, MacGregor, Morey, Whitney, Brenner, Steigerwalt, Helsel, Raney, Patel, Levett and Brown 2005, 7 (Effective publication: Daneshvar, Hollis, Weyant, Jordan, MacGregor, Morey, Whitney, Brenner, Steigerwalt, Helsel, Raney, Patel, Levett and Brown 2004, 4197.)
au.ri.mu.co'sum. L. n. aurum gold; L. neut. adj. mucosum slimy; N.L. neut. adj. aurimucosum slimy and gold-colored, pertaining to the appearance of colonies.
Cells are Gram-stain-positive and non-acid-fast. They are thin, nonmotile, nonsporeforming, and pleomorphic coryneforms. On Columbia blood agar supplemented with 5% sheep blood, the colonies are sticky and slightly yellow or charcoal black in color. On trypticase soy agar, they appear colorless or charcoal black and slimy. On brain heart infusion agar supplemented with 1% Tween 80, some strains are able to form a corraloid precipitin in agar when grown in the presence of CO2. Facultatively anaerobic. Oxidase very rarely positive. Acid is produced from fructose, D-glucose, maltose, sucrose, and occasionally D-mannitol but not from adonitol, amygdalin, arabinose, cellobiose, glycerol, glycogen, inulin, lactose, mannose, melezitose, raffinose, rhamnose, ribose, salicin, sorbitol, trehalose, or xylose. Hydrolyzes hippurate and occasionally gelatin and urea but not starch. Esculin hydrolysis is variable, and nitrate reductase is rare. Positive for alkaline phosphatase, leucine arylamidase, and pyrazinamidase activities, but negative for acid phosphatase, arginine dihydrolyse, esterase, ester lipase, lipase, α-glucosidase, β-glucosidase, α-galactosidase, β-galactosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, α-mannosidase, α-fucosidase, phosphoamidase, pyrrolidonyl arylamidase, valine arylamidase, cystine arylamidase, trypsin, and chymotrypsin. Acetoin positive and indole negative. Lactate, not propionate, is the major end product of glucose fermentation.
Cells contain meso-DAP with galactose and arabinose sugars in whole-cell hydrolysates (i.e. the cell-wall chemotype is chemotype IV). Short-chain corynomycolic acids are present. The fatty acid profile contains saturated, unsaturated, and tuberculostearic acids. It has type PI phospholopid pattern with no nitrogen-containing compounds.
Source: human clinical specimens; black-pigmented colony types are from female urogenital sites, some of which are associated with complications in pregnancy.
DNA G+C content (mol%): 63.7 (HPLC); 60.6 by complete genome sequence analysis of strain CN-1.
Type strain: CCUG 47449, DSM 44532, IMMIB D-1488, JCM 11766, NRRL B-24143.
Sequence accession no. (16S rRNA gene): AJ309207.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492282. Corynebacterium nigricans (Shukla et al. 2004) (Effective publication: Shukla et al. 2003.) was reassigned as a later heterotypic synonym of Corynebacterium aurimucosum (Yassin et al. 2002a), Daneshvar 2005 (Effective publication: Daneshvar et al. 2004.). The complete genome of a black-pigmented Corynebacterium aurimucosum strain CN-1 (= ATCC 700975, type strain of Corynebacterium nigricans) with 2,790,189 bp has been deposited under GenBank accession no. CP001601 (Trost et al., 2010). By 16S rRNA gene sequencing alone, this species can not be readily discerned from Corynebacterium minutissimum and Corynebacterium singulare, but these species can be resolved using rpoB gene sequencing (Khamis et al., 2004).
Corynebacterium auris
Funke, Lawson and Collins 1995c, 738VP
au'ris. L. gen. fem. n. auris of the ear.
Gram-stain-positive, nonmotile, nonsporeforming diphtheroids. Colonies are circular, convex, and dry, becoming slightly yellowish with time; slightly adhere to agar. Weakly adherent if subcultured on agar plates. Nonlipophilic. Oxidative metabolism. Acid is not produced from glucose, maltose, sucrose, mannitol, xylose, ribose, lactose, and glycogen. The following substrates are utilized: γ-hydroxybutyric acid, L-malic acid, pyruvic acid, succinamic acid, N-acetyl-L-glutamic acid, L-asparagine, L-glutamic acid, glycyl-L-glutamic acid, and L-pyroglutamic acid. Nitrate is not reduced. Urea and esculin are not hydrolyzed. The CAMP reaction is positive. Pyrazinamidase, alkaline and acid phosphatases, esterase, esterase lipase, lipase, leucine arylamidase, and phosphoamidase are produced.
The cell wall contains meso-DAP. Mycolic acids are produced but are cleaved at the temperature (300°C) produced in the injection port of the commercial MIDI system, resulting in fatty acids which are identified as, e.g. C17:1 ω6c–C17:1 ω9c (Funke and Bernard, 2007). Cellular fatty acids are consistent with those described for the genus. Propionic acid not detected as metabolic product (Bernard et al., 2002).
Source: outer ear channel of humans.
DNA G+C content (mol%): 68–74 (HPLC).
Type strain: strain ATCC 51966, CCUG 33426, CIP 104632, DMMZ 328, DSM 44122, JCM 11946, LMG 19072.
Sequence accession no. (16S rRNA gene): X81873.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492234. Corynebacterium auris was originally described as Coryneform CDC group ANF-1 like.
Corynebacterium auriscanis
Collins, Hoyles, Lawson, Falsen, Robson and Foster 2000, 423VP (Effective publication: Collins, Hoyles, Lawson, Falsen, Robson and Foster 1999b, 3446.)
au.ris.ca'nis. L. fem. n. auris ear; L. masc. n. canis dog; N.L. gen. n. auriscanis of the ear of the dog.
Cells are Gram-stain-positive typically club-shaped rods, which appear as single cells, in pairs, or in clusters. Obligately aerobic. Nonlipophilic. CAMP reaction negative. Acid is produced from glucose but not from glycogen, lactose, maltose, mannitol, sucrose, ribose, or D-xylose. Nitrate is not reduced, and the Voges–Proskauer test is negative. Hippurate is hydrolyzed. Esculin hydrolysis is variable. Urea, gelatin, and starch are not hydrolyzed. Alkaline and acid phosphatases, esterase (weak reaction), ester lipase (weak reaction), leucine arylamidase, phosphoamidase, and pyrrolidonyl arylamidase activities are detected. N-Acetyl-β-glucosaminidase, chymotrypsin, cystine arylamidase, α-fucosidase, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, β-glucuronidase, α-mannosidase, pyrazinamidase, trypsin, and valine arylamidase activities are not detected. Lipase activity variable.
The cell wall contains meso-DAP. Short-chain mycolic acids (C28–C34) are present, with C30:0, C32:0, and C34:0 as the main components. Cellular fatty acids are consistent with those described for the genus, but tuberculostearic acid is not present.
Source: clinical specimens from dogs; habitat is not known.
DNA G+C content (mol%): 61 (Tm).
Type strain: M598/96/1, CCUG 39938, CIP 106629, DSM 44609, JCM 12369.
Sequence accession no. (16S rRNA gene): AJ243819.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492244.
Corynebacterium bovis
Bergey, Harrison, Breed, Hammer and Huntoon 1923, 388AL
bo'vis. L. n. bos a cow; L. gen. n. bovis of a cow.
(This description is largely based on that of Collins and Cummins, 1986.)
Irregular rods 0.5–0.7 × 2.5–3.0 µm, often clubbed shaped; coccobacillary forms may occur. Colonies on nutrient agar supplemented with Tween 80 (0.1%) are white to cream, circular, entire, slightly shiny, ∼1–2 mm in diameter. Aerobic. Lipophilic. Most strains ferment glucose, fructose, galactose, maltose, and glycerol. Oxidase, arabinose, lactose, trehalose, and dextrin variable. Xylose, rhamose, lactose, mannitol, mannose, sucrose, raffinose, salicin, and starch not utilized. Esculin, starch, and casein not hydrolyzed. Hippurate hydrolyzed; phosphatase and pyrazinamidase positive, but variable for β-galactosidase. Nitrate not reduced; urease not produced (Collins and Cummins, 1986; Hollis and Weaver, 1981).
The cell wall contains meso-DAP (variation A1γ), with sugars arabinose and galactose. Low-molecular-weight mycolic acids (∼C22–C36) found, with the α-alkyl group side-chain consisting of C6H13 and C8H17. MK-9 (H2) detected. Cellular fatty acids are consistent with those described for the genus (Bernard et al., 1991), with TBSA being detected. Propionic acid not detected as metabolic product (Bernard et al., 2002).
Source: aseptically drawn milk, as a commensal on cow's udder or as cause of bovine mastitis and occasionally as cause of infection in humans (Achermann et al., 2009; Bernard et al., 2002; Dutly et al., 2003; Vale and Scott, 1977).
DNA G+C content (mol%): 67.8–69.7 (Tm).
Type strain: ATCC 7715, CCUG 2705, CIP 54.80, DSM 20582, JCM 11947, NCTC 3224.
Sequence accession no. (16S rRNA gene): X84444.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492236.
Corynebacterium callunae
(Lee and Good, 1963) Yamada and Komagata 1972, 412AL (“Corynebacterium callunae” Lee and Good 1963, 1349.)
cal.lu'na.e. N.L. n. calluna generic name of heather; N.L. gen. n. callunae of heather.
(This description is largely based on that of Collins and Cummins, 1986.)
Short, strongly Gram-stain-positive rods, metachromatic granules present. Moderate growth on nutrient agar. Glucose, fructose, mannose, maltose, sucrose, trehalose, salicin, and methyl red positive. Hippurate hydrolyzed and urease produced. Negative for arabinose, xylose, rhamnose, galactose, lactose, raffinose, dextrin, and starch fermentation. Esculin, casein, and gelatin hydrolysis negative, and nitrate not reduced. Grows at pH 6 and in 30% glucose but not at 45°C (Fudou et al., 2002).
Cell wall contains meso-DAP. Arabinose and galactose are major sugars. Mycolates are present. Cellular fatty acids are consistent with those of the genus. Menaquinone is MK-9(H2).
Source: heather; habitat remains unknown.
DNA G+C content (mol%): 51.2 (Tm).
Type strain: ATCC 15991, CCUG 28793, CIP 104277, DSM 20147, HAMBI 2053, JCM 9489, NBRC 15359.
Sequence accession no. (16S rRNA gene): X84251.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492245.
Corynebacterium camporealensis
Fernández-Garayzábal, Collins, Hutson, Gonzalez, Fernández and Domínguez 1998, 466VP
cam.po.re.al.en'sis. N.L. adj. camporealensis pertaining to Campo Real, Madrid, Spain.
Gram-stain-positive pleomorphic rods occurring singly or are arranged in palisades or V-shaped forms. Colonies are nonhemolytic, circular, slightly convex, smooth, and with a creamy consistency. Facultatively anaerobic. Nonlipophilic. CAMP reaction is strongly positive. Nitrate is not reduced. Esculin, urea, and gelatin are not hydrolyzed. Acetoin and indole are not produced. Acid is produced from glucose, but not from ribose, xylose, mannitol, lactose, maltose, sucrose, and glycogen. Pyrazinamidase, alkaline phosphatase, esterase, esterase lipase, lipase, leucine arylamidase, valine arylamidase, and cystine arylamidase are produced. Pyrrolidonyl arylamidase, acid phosphatase, β-glucuronidase, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, α-mannosoidase, α-fucosidase, N-acetyl-β-glucosaminidase, trypsin, and chymotrypsin are not produced.
The cell wall contains meso-diaminopimelic acid. Short-chain mycolic acids are present.
Source: milk of sheep affected with subclinical mastitis.
DNA G+C content (mol%): not available.
Type strain: CRS-51, ATCC BAA-77, CCUG 39412, CECT 4897, CIP 105508, DSM 44610, JCM 11664.
Sequence accession no. (16S rRNA gene): Y09569.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492246.
Corynebacterium canis
Funke, Englert, Frodl, Bernard and Stenger 2010a, 2546VP
ca'nis. L. gen. n. canis of a dog.
Cells are Gram-stain-positive, nonsporeforming, and non-motile. Some cells are typically club-shaped, rods but some are filamentous, and some even show branching. Colonies are beige-whitish, dryish, with irregular edges, convoluted, up to 1–2 mm in diameter after 48 h incubation, and adherent to sheep blood agar. Facultatively anaerobic. Catalase positive. Acid is produced from glycerol, galactose, glucose, fructose, mannose, arbutin, salicin, maltose, sucrose, trehalose, starch, glycogen, tagatose, and 5-keto-gluconate but not from erythritol, arabinose, ribose, xylose, adonitol, methyl-β-D-xylopyranoside, L-sorbose, inositol, D-mannitol, D-sorbitol, methyl-α-D-mannopyranoside, methyl-α-D-glucopyranoside, N-acetylglucosamine, amygdalin, D-cellobiose, D-lactose, D-melibiose, inulin, D-melezitose, xylitol, gentiobiose, D-turanose, D-lyxose, fucose, arabitol, gluconate, and 2-keto-gluconate. Activities of the following enzymes can be detected: nitrate reductase, pyrazinamidase, β-glucosidase, α-glucosidase, alkaline phosphatase, esterase, esterase lipase, leucine arylamidase, trypsin, and phosphoamidase. Activities of urease, pyrrolidonyl arylamidase, gelatinase, lipase, cystine arylamidase, α-chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase are not detected. The CAMP reaction is negative.
The cell wall contains meso-DAP. Mycolic acids are present. Cellular fatty acids are consistent with those described for the genus.
Source: a patient's wound after a dog bite.
DNA G+C content (mol%): not available.
Type strain: 1170, CCUG 58627, DSM 45402.
Sequence accession no. (16S rRNA gene): GQ871934.
Further comments: additional GenBank accession numbers include (partial rpoB gene) GQ871935.
Corynebacterium capitovis
Collins, Hoyles, Foster, Sjödén and Falsen 2001a, 858VP
ca.pit.o'vis. L. gen. n. capitis of a head; L. gen. n. ovis of a sheep; N.L. gen. n. capitovis of a sheep's head.
Cells are Gram-stain-positive diphtheroids. Colonies are circular, entire, convex, nonhemolytic, and lemon-pigmented. Facultatively anaerobic. Nonlipophilic. Acid is produced from glucose but not from lactose, maltose, mannitol, ribose, sucrose, and D-xylose. Nitrate is not reduced. Esculin, urea, and gelatin are not hydrolyzed. Activity for alkaline and acid phosphatases, esterase (weak), esterase lipase, lipase (weak), and leucine arylamidase is detected. No activity is detected for cystine arylamidase, chymotrypsin, α-fucosidase, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, β-glucuronidase, α-mannosidase, N-acetyl-β-glucosaminidase, valine arylamidase, pyrazinamidase, phosphoamidase, pyrrolidonyl arylamidase, and trypsin.
Cell wall contains meso-DAP. Long-chain fatty acids are of the straight-chain saturated and monounsaturated types. Tuberculostearic acid is not present. Mycolic acids are present (C32–C36).
Source: infected head of a sheep; habitat is unknown.
DNA G+C content (mol%): not available.
Type strain: S108/98/2, CCUG 39779, CIP 106739, DSM 44611, JCM 12101.
Sequence accession no. (16S rRNA gene): AJ297402.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492247.
Corynebacterium casei
Brennan, Brown, Goodfellow, Ward, Beresford, Simpson, Fox and Cogan 2001, 850VP
ca'se.i. L. gen. n. casei of cheese, named because the organism was isolated from cheese.
Cells are Gram-stain-positive irregularly shaped rods of 1–3 µm in length. Colonies are cream, circular, matt, entire, low-concave, and 1 mm in diameter. Facultatively anaerobic. Nitrate is reduced to nitrite. Esculin, urea, gelatin, tyrosine, and ONPG are not hydrolyzed. CAMP reaction is negative. Acid is produced from glucose, ribose, mannose, and fructose but not from glycogen, glycerol, erythritol, D-arabinose, L-arabinose, D-xylose, L-xylose, adonitol, methyl-β-xyloside, galactose, sorbose, rhamnose, dulcitol, inositol, mannitol, sorbitol, methyl-α-D-mannoside, methyl-α-D-glucoside, amygdalin, arbutin, salicin, cellobiose, maltose, lactose, melibiose, sucrose, inulin, melezitose, raffinose, xylitol, gentiobiose, D-turanose, D-lyxose, D-tagatose, D-fucose, L-fucose, D-arabitol, L-arabitol, gluconate, or 2-ketogluconate. Alkaline and acid phosphatases, esterase, esterase lipase, leucine arylamidase, cystine arylamidase, and pyrazinamidase activities are present. The isolates do not produce lipase, trypsin, chymotrypsin, α-galactosidase, α-mannosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, and N-acetyl-β-glucosaminidase.
The cell wall was found to be type IV, containing meso-diaminopimelic acid, arabinose, galactose, and short-chain mycolic acids (C22–C36). The menaquinones in the cell wall were of the MK-9(H2) type. Analysis of the polar lipid of the cell wall revealed the presence of diphosphatidylglycerol, phosphatidylglycerol, phosphotidylinositol, and phosphatidylglycerol mannosides.
Source: surface of smear-ripened cheese.
DNA G+C content (mol%): 51 (HPLC).
Type strain: CIP 107182, DPC 5298, DSM 44701, JCM 12072, LMG S-19264, NCIMB 30130.
Sequence accession no. (16S rRNA gene): AF267152.
Further comments: additional GenBank accession numbers include (complete rpoB gene) EU616817.
Corynebacterium caspium
Collins, Hoyles, Foster and Falsen 2004, 926VP
cas.pi'um. L. neut. adj. caspium belonging to the Caspian Sea, referring to the isolation of the type strain from a Caspian seal.
Cells are Gram-stain-positive, irregular-shaped tapered rods and clubs. Nonlipophilic. Colonies are circular, convex, entire, opaque, dull, nonhemolytic and can be moved across the plate while retaining their integrity. Facultatively anaerobic. Nitrate is not reduced. CAMP reaction negative. Acid is produced from D-glucose and D-ribose, but not from glycogen, lactose, maltose, mannitol, sucrose, or D-xylose. Urea is hydrolyzed but esculin and gelatin are not. Activity is detected for esterase, esterase lipase, pyrazinamidase, and trypsin. No activity is detected for acid phosphatase, alkaline phosphatase, chymotrypsin, cystine arylamidase, α-fucosidase, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, lipase, leucine arylamidase, α-mannosidase, pyrrolidonyl arylamidase, phosphoamidase, or valine arylamidase.
Cell-wall murein is based on meso-diaminopimelic acid. Corynomycolic acids are not present. Cellular fatty acids are consistent with those described for the genus; tuberculostearic acid is not present.
Source: a Caspian seal; habitat is not known.
DNA G+C content (mol%): not available.
Type strain: M/106/00/5, CCUG 44566, CIP 107965, JCM 13387.
Sequence accession no. (16S rRNA gene): AJ566641.
Corynebacterium ciconiae
Fernández-Garayzábal, Vela, Egido, Hutson, Lanzarot, Fernández-García and Collins 2004, 2194VP
ci.co.ni'a.e. L. gen. fem. n. ciconiae of a (black) stork.
Cells are Gram-stain-positive rods. Nonlipophilic. Colonies are creamy-white, circular, convex, dry, and nonhemolytic on Columbia blood agar. Facultatively anaerobic. CAMP reaction negative. Nitrate is not reduced. Esculin, urea, and gelatin are not hydrolyzed. Acid is produced from glucose, ribose, glycerol, D-fructose, D-mannose, N-acetyl-β-glucosamine, and maltose, but not from D-xylose, mannitol, lactose, sucrose, glycogen, erythritol, L-arabinose, L-xylose, adonitol, methyl β-xyloside, galactose, sorbose, L-rhamnose, dulcitol, inositol, sorbitol, methyl α-D-mannoside, methyl α-D-glucoside, amygdalin, arbutin, salicin, cellobiose, melibiose, inulin, melezitose, D-raffinose, xylitol, b-gentiobiose, D-turanose, D-lyxose, D-tagatose, D-fucose, D-arabitol, L-arabitol, 2-keto-gluconate, or 5-keto-gluconate. Acidification of D-arabinose, L-fucose, trehalose, starch, and gentiobiose is variable. Activity of alkaline and acid phosphatases, esterase, ester lipase, lipase, leucine arylamidase, phosphoamidase, β-glucosidase, and pyrazinamidase is detected. Pyrrolidonyl arylamidase, β-glucuronidase, β-galactosidase, α-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, α-galactosidase, α-fucosidase, chymotrypsin, trypsin, valine arylamidase, and cystine arylamidase are not produced. Mycolic acids are absent. Cellular fatty acids are consistent with those described for the genus.
Source: the trachea of apparently healthy wild nesting black storks (Ciconia nigra).
DNA G+C content (mol%): not available.
Type strain: BS13, CCUG 47525, CECT 5779, JCM 13388.
Sequence accession no. (16S rRNA gene): AJ555193.
Corynebacterium confusum
Funke, Osorio, Frei, Riegel and Collins 1998c, 1294VP
con.fu'sum. L. past part. confusum confused, to indicate that this bacterium might be phenotypically confused with many other Corynebacterium species.
Cells are Gram-stain-positive, typically occurring as club-shaped rods as single cells, in pairs, or in small clusters. Colonies are whitish, glistening, convex, and creamy, up to 1.5 mm in diameter after 48 h incubation. Facultatively anaerobic, but weak anaerobic growth. Nonlipophilic. CAMP reaction is negative. Nitrate is reduced. Esculin, urea, and tyrosine are not hydrolyzed. Acid is produced from D-glucose, ribose, D-fructose, tagatose, and 5-ketogluconate, but acid is not produced from maltose, sucrose, mannitol, D-xylose, glycerol, erythritol, arabinose, adonitol, β-methylxyloside, galactose, L-sorbose, rhamnose, dulcitol, inositol, sorbitol, methyl-α-D-mannoside, methyl-α-D-glucoside, N-acetylglucosamine, amygdalin, arbutin, salicin, cellobiose, lactose, melibiose, trehalose, inulin, melezitose, D-raffinose, glycogen, xylitol, D-turanose, D-lyxose, fucose, arabitol, gluconate, or 2-ketogluconate. Acid production from β-gentiobiose is variable. Activities of pyrazinamidase, alkaline phosphatase, esterase, and esterase lipase are detected, but pyrrolidonyl arylamidase, valine arylamidase, trypsin, chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase are not detected. Activities of leucine arylamidase and phosphoamidase are variable.
The cell wall contains meso-diaminopimelic acid. Mycolic acids are present. Main straight-chain saturated fatty acids are palmitic and stearic acids; oleic acid is the predominant unsaturated fatty acid. Minor amounts of tuberculostearic acid are present. A clinical strain was CAMP positive, and propionic acid was detected as a metabolic product (Bernard et al., 2002).
Source: human clinical specimens.
DNA G+C content (mol%): not available.
Type strain: CCUG 38267, CIP 105403, DMMZ 2439, DSM 44384, JCM 12102.
Sequence accession no. (16S rRNA gene): Y15886.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492248.
Corynebacterium coyleae
Funke, Pascual Ramos and Collins 1997e, 94VP
coy.le'a.e. N.L. gen. fem. n. coyleae of Coyle, to honor the American microbiologist Marie B. Coyle for her contributions to the clinical microbiology of coryneform bacteria.
Cells are Gram-stain-positive diphtheroids. Colonies are whitish, circular, convex, and slightly glistening; some colonies have a creamy consistency, and others have a sticky consistency and are ∼1 mm after 24 h incubation at 37°C on blood agar. Facultatively anaerobic, but very weak anaerobic growth. Nonlipophilic. Acid is slowly produced from glucose, ribose, D-fructose, D-mannose, and 5-keto-gluconate but not from maltose, sucrose, mannitol, xylose, glycerol, erythritol, D-arabinose, L-arabinose, adonitol, β-methylxyloside, galactose, L-sorbose, rhamnose, dulcitol, inositol, sorbitol, α-methyl-D-mannoside, α-methyl-D-glucoside, N-acetylglucosamine, amygdalin, arbutin, salicin, cellobiose, lactose, melibiose, trehalose, inulin, melezitose, D-raffinose, starch, glycogen, xylitol, b-gentiobiose, D-turanose, D-lyxose, D-tagatose, D-fucose, L-fucose, D-arabitol, L-arabitol, gluconate, and 2-keto-gluconate. Nitrate is not reduced. Urea, esculin, casein, tyrosin, and xanthine are not hydrolyzed. The CAMP reaction is strongly positive. Pyrazinamidase, alkaline and acid phosphatases, esterase, esterase lipae, leucine aryamidase, cystine arylamidase, and phosphoamidase activities are detected, but valine arylamidase, chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase activities are not detected. Pyrrolidonyl arylamidase activity is variable.
The cell wall contains meso-DAP. Mycolic acids are present. Cellular fatty acids are consistent with those described for the genus. Does not produce propionic acid as metabolic product (Bernard et al., 2002).
Source: human clinical specimens.
DNA G+C content (mol%): 62–64 (Tm).
Type strain: ATCC 700219, CCUG 38194, CIP 104919, DMMZ 214, DSM 44184, JCM 10381.
Sequence accession no. (16S rRNA gene): X96497.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492250. Six Corynebacterium coyleae strains associated with neonatal bacteremias were found to possess ermX and express resistance to MLS antibiotics (Fernandez-Natal et al., 2008). By 16S rRNA gene sequencing, Corynebacterium coyleae is closely related to (98% identity with) Corynebacterium mucifaciens and Corynebacterium afermentans but differs from them (∼94% identity with) when rpoB gene sequences are compared (Khamis et al., 2004) as well as by phenotypic means (Table 2).
Corynebacterium cystitidis
Yanagawa and Honda 1978, 215AL
cys.ti'ti.dis. Gr. n. kustis bladder; N.L. n. cystitis cystitis; N.L. gen. n. cystitidis of cystitis.
(This description is largely based on that of Collins and Cummins, 1986.)
Gram-stain-positive rods, 0.5–1.3 µm singly, in pairs often at angles, or in irregular masses. Fimbriae (pili) visible using electron microscopy. Colonies on nutrient agar and serum agar are creamy to pale yellow, entire, circular, opaque, and 1 mm in diameter at 24 h. No hemolysis on various blood agars. Pellicle and granular sediment formed in broth. No growth at 5°C; grows at 41.5°C, and cells remain viable for 30 min at 56°C. Aerobic and facultatively anaerobic. Complex vitamin and amino acid requirements. Positive for glucose, xylose, fructose, maltose, trehalose, dextrin, and starch; negative for arabinose, rhamnose, galactose, mannose, lactose, sucrose, raffinose, and salicin. Nitrate is not reduced. Urease is produced, hippurate and starch are hydrolyzed, and pyrazinamidase is detected. Tyrosine, esculin, and gelatin are not hydrolyzed; casein is not digested; methyl red, and phosphatase are negative.
Meso-DAP and sugars arabinose and galactose are present. MK-8(H2) detected.
Source: cows with severe hemorrhagic cystitidis and from the prepuce of healthy bulls.
DNA G+C content (mol%): 52.6–53.9 (Tm).
Type strain: ATCC 29593, CCUG 28794, CIP 103424, DSM 20524, JCM 3715, NBRC 15284, NCTC 11863.
Sequence accession no. (16S rRNA gene): X84252.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492251.
Corynebacterium doosanense
Lee, Cho, Jung, Van Nguyen, Jung, Park, Le and Kim 2009, 2736VP
do.o.san.en'se. N.L. neut. adj. doosanense belonging to Doosan, named after the Doosan group, a foundation of Chung-Ang University, Seoul, Korea where taxonomic studies on this species were performed.
Gram-stain-positive, nonmotile, nonsporeforming irregular and club-shaped rods that are 0.8∼1.0 × 1.0∼1.2 µm in size. Colonies are 1∼2 mm in diameter, yellow, low convex on GYEA media, and yellow, opaque, low-convex on sheep blood agar after 48 h incubation at 30°C. Aerobic. Activities for sodium pyruvate, pyrazinamidase, ester lipase, leucine arylamidase, and naphthol-AS-BI-phosphohydrolase are detected. Reduction of nitrate and hippurate hydrolysis are positive. No activity is detected for pyrrolidonyl arylamidase, alkaline phosphatase, esterase, lipase, valine arylamidase, cystine arylamidase, trypsin, acid phospatase, α-chymotrypsin, β-glucuronidase, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, α-mannosidase, α-fucosidase, N-acetyl-β-glucosaminidase, leucine aminopeptidase, urease, gelatin, and esculin hydrolysis. Acid production occurs only from glucose.
The cell wall contains meso-DAP. Whole cell hydrolysates contain mainly galactose and arabinose. Mycolic acids are detected. Cellular fatty acids are consistent with those described for the genus.
Source: activated sludge, taken from a wastewater treatment plant in Yeongdeuk-gun, Republic of Korea.
DNA G+C content (mol%): 53.5 (Tm).
Type strain: CAU 212, CCUG 57284, KCTC 19568.
Sequence accession no. (16S rRNA gene): EU998655.
Corynebacterium durum
Riegel, Heller, Prévost, Jehl and Monteil 1997a, 1110VP
du'rum. L. neut. adj. durum hard, tough.
The bacteria are Gram-stain-positive pleomorphic rods which can be long and sometimes filamentous. After aerobic incubation at 37°C for 72 h, colonies are beige, convex, and rough, with convolutions and an irregular margin, and strongly adhere to the agar. Colonies incubated in an aerobic atmosphere supplemented with 10% CO2 have a dense center and nearly regular margins; they adhere weakly to the agar. Formation of filaments under these conditions is rare. Facultatively anaerobic, but weak anaerobic growth. Nitrate reduction positive, but urea hydrolysis is variable. A weak hydrolysis of esculin can be observed sometimes after at least 72 h of incubation. Gelatin and tyrosine are not degraded. Acid is produced from glucose, maltose, fructose, sucrose, galactose, and mannitol but not from ribose, lactose, trehalose, glycogen, or xylose. Pyrazinamidase is produced, but pyrrolidonyl arylamidase, alkaline phosphatase, β-glucuronidase, β-galactosidase, α-glucosidase, and N-acetyl-β-glucosaminidase are not. Propionic acid, but not succinic acid is produced as end product of anaerobic metabolism of glucose.
The cell wall contains meso-DAP, with sugars arabinose and galactose. Mycolic acids with short chain lengths (C26–C36) are present (HPLC method). Cellular fatty acids are consistent with those described for the genus (Rassoulian-Barrett et al., 2001).
Source: human respiratory tract specimens and occasionally other normally sterile sites.
DNA G+C content (mol%): 55 (capillary electrophoresis).
Type strain: IBS G15036, CCUG 37331, CIP 105490, DSM 44351, JCM 11948.
Sequence accession no. (16S rRNA gene): Z97069.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492252. Corynebacterium durum is the most prominent Corynebacterium recovered from throat swabs of healthy adults (von Graevenitz et al., 1998).
Corynebacterium efficiens
Fudou, Jojima, Seto, Yamada, Kimra, Nakamatsu, Hiraishi and Yamanaka 2002, 1130VP
ef.fi.ci'ens. L. part. adj. efficiens effecting, effective, efficient.
Cells are Gram-stain-positive club-shaped rods 0.8–1.1 × 1.0–4.5 µm in size. Colonies on nutrient agar are smooth, entire, circular, dull to slightly glistening, and generally yellow. Facultatively anaerobic. All strains require biotin for growth. Good growth at 30–40°C; growth occurs up to 45°C. No growth occurs when the pH is below 6 or in the presence of 30% glucose. Nitrate is reduced to nitrite. Hydrolysis of esculin and urea is variable. Acid is formed from glucose, fructose, mannose, ribose, maltose, and dextrin, but not from xylose, mannitol, lactose, salicin, galactose, starch, or glycogen. All strains assimilate acetate, pyruvate, and L-lactate, but they do not assimilate D-lactate, succinate, 2-oxoglutarate, citrate, adipicate, pimelate, glycolate, or glyoxylate. Pyrazinamidase is detected but pyrrolidonyl arylamidase, alkaline phosphatase, β-galactosidase, α-glucosidase, N-acetyl-β-glucosaminidase, and tyrosinase are not detected. Produces large amounts of L-glutamic acid under aerobic conditions.
The cell wall contains meso-DAP. Mycolic acids are present. MK-9(H2) is the major menaquinone. Cellular fatty acids are consistent with those described for the genus. Polar lipids include phosphatidylinositol and its mannoside.
Source: soil and vegetables.
DNA G+C content (mol%): 59–60.2 (Tm), 63.4 (complete genome sequencing).
Type strain: YS-314, AJ 12310, CCUG 47130, CCUG 48037, DSM 44549, JCM 11189, NBRC 100395.
Sequence accession no. (16S rRNA gene): AB055963.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AP005215. The complete genome of Corynebacterium efficiens YS-314T, deposited under GenBank accession no. NC_004369 with 3,147,090 bp, as well as discussion of how amino acid substitutions positively affect thermostability, have been reported (Brune et al., 2005; Nishio et al., 2003).
Corynebacterium falsenii
Sjödén, Funke, Izquierdo, Akervall and Collins 1998, 73VP
fal.se'ni.i. N.L. gen. masc. n. falsenii of Falsen, to honor the Swedish microbiologist and taxonomist Enevold Falsen curator of the CCUG, for his lifelong contributions to bacterial taxonomy as well as for the systematic collection and characterization of prokaryotes.
Gram-stain-positive, typically club-shaped rods which appear as single cells, pairs, or in small clusters. Nonlipophilic. Colonies are whitish, circular with entire edges, convex, glistening, of creamy consistency, and start to develop a yellowish pigment after 72 h incubation. Weak anaerobic growth. Acid is produced from glucose, galactose, 5-keto-gluconate, glycerol, ribose, and trehalose, but not from sucrose, mannitol, xylose, adonitol, amygdalin, arabinose, arabitol, arbutin, cellobiose, dulcitol, erythritol, D-fucose, β-gentiobiose, gluconate, 2-keto-gluconate, N-acetylglucosamine, methyl α-D-glucoside, glycogen, inositol, inulin, D-lyxose, methyl α-D-mannoside, melezitose, melibiose, D-raffinose, salicin, sorbitol, sorbose, D-turanose, xylitol, and β-methyl-xyloside. Acid production from maltose, fructose, lactose, rhamnose, and D-tagatose is variable. Nitrate is not reduced. Urea hydrolysis is positive but delayed. Esculin, casein, tyrosine, and xanthine are not hydrolyzed. The CAMP reaction is negative. Pyrazinamidase, alkaline and acid phosphatase, esterase, esterase lipase, and phosphoamidase activities are detected, but pyrrolidonyl arylamidase, valine arylamidase, trypsin, chymotrypsin, α-galactosidase, β-glucosidase, N-acetylglucosaminidase, α-mannosidase, and α-fucosidase activities are not detected. Lipase, leucine arylamidase, and cystine arylamidase activities are variable.
The cell wall contains meso-DAP. Mycolic acids are present. Cellular fatty acids are consistent with those described for the genus. Does not produce propionic acid as metabolic product (Bernard et al., 2002).
Source: human clinical specimens.
DNA G+C content (mol%): not available.
Type strain: Y13024, CCUG 33651, CIP 105466, DSM 44353, JCM 11949.
Sequence accession no. (16S rRNA gene): Y13024.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492253. Corynebacterium falsenii was previously known as EF (Enevold Falsen) group 42 bacteria.
Corynebacterium felinum
Collins, Hoyles, Hutson, Foster and Falsen 2001b, 1351VP
fe.li'num. L. neut. adj. felinum pertaining to cats.
Cells are Gram-stain-positive diphtheroids. Colonies are nonhemolytic. Facultatively anaerobic. Nonlipophilic. Acid is produced from glucose, maltose, and ribose but not from lactose, mannitol, sucrose, N-acetyl-β-glucosamine, and D-xylose. Activity is detected for α-glucosidase, pyrazinamidase, pyrrolidonyl arylamidase, and leucine arylamidase. No activity is observed for alkaline and acid phosphatases, chymotrypsin, esterase, ester lipase, lipase, α-galactosidase, β-galactosidase, β-glucosidase, β-glucuronidase, α-fucosidase, α-mannosidase, phosphoamidase, valine arylamidase, and trypsin. Nitrate is not reduced. Esculin, urea, and gelatin are not hydrolyzed.
The cell wall contains meso-DAP. Mycolic acids (C32–C36) detected. MK-8(H2) is the predominant menaquinone. Cellular fatty acids are consistent with those described for the genus, but tuberculostearic acid is not present.
Source: a dead Scottish wild cat (Felis sylvestris); habitat is unknown.
DNA G+C content (mol%): not available.
Type strain: M714/95/5, CCUG 39943, CIP 106740, DSM 44508, JCM 12103.
Sequence accession no. (16S rRNA gene): AJ401282.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492254.
Corynebacterium flavescens
Barksdale, Lanéelle, Pollice, Asselineau, Welby and Norgard 1979, 222AL (Microbacterium flavum Orla-Jensen 1919, 181)
fla.ves'cens. L. v. flavescere to become yellow; L. part. adj. flavescens becoming yellow.
(This description is largely based on that of Collins and Cummins, 1986.)
Gram-stain-positive to Gram-stain-variable pleomorphic rods, often with tapered ends, showing metachromatic granules. Growth is yellow on Loeffler's agar; on BHI agar, colonies are smooth, butyrous, and cream-colored. Colonies on tellurite agar are grayish black, later developing grayish white centers. Aerobic and facultatively anaerobic. Complex nutritional requirements.
Glucose, fructose, galactose, mannose, and methyl red are positive. Negative for arabinose, xylose, rhamnose, lactose, maltose, sucrose, trehalose, raffinose, salicin, dextrin, and starch fermentation. Hippurate and gelatin hydrolysis negative. Nitrate not reduced, urease not detected, and pyrazinamidase negative. Reactions for esculin, tyrosine, and casein hydrolysis unknown.
Information regarding DAP species, menaquinones, presence of mycolates unknown. Cellular fatty acids are consistent with those of the genus (Bernard et al., 1991).
Source: dairy products.
DNA G+C content (mol%): 58.3 (Tm).
Type strain: 8 of Orla-Jensen, ATCC 10340, CCUG 28791, CIP 69.5, DSM 20296, JCM 1317, LMG 4046, NBRC 14136, NCCB 42012, NCIMB 8707, VKM Ac-1956.
Sequence accession no. (16S rRNA gene): X84441.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492255.
Corynebacterium freiburgense
Funke, Frodl, Bernard and Englert 2009, 2056VP
frei.bur.gen'se. N.L. neut. adj. freiburgense from Freiburg/Breisgau, Germany, named after the city where the bacterium was first isolated.
Gram-stain-positive typically club-shaped rods that occur as single cells, in pairs, or in small clusters. Colonies are beige-whitish, dryish, with irregular edges, adherent to agar, convoluted, and up to 1–2 mm in diameter after 48 h. In 5-d-old colonies, a “spoke-wheel” macroscopic morphology may be observed. Facultatively anaerobic, but anaerobic growth is weak. Nonlipophilic. Acid is produced from glucose, maltose, sucrose, fructose, galactose, 5-keto-gluconate, lactose, mannose, ribose, and tagatose, but acid is not produced from N-acetylglucosamine, adonitol, amygdalin, arabinose, arabitol, arbutin, cellobiose, dulcitol, erythritol, fucose, gentiobiose, gluconate, glycerol, inositol, inulin, 2-keto-gluconate, lyxose, mannitol, melezitose, melibiose, methyl-α-D-glucoside, methyl-α-D-mannoside, β-methyl-xyloside, D-raffinose, rhamnose, salicin, sorbitol, sorbose, starch, trehalose, D-turanose, xylitol, and xylose. Activity of the following enzymes can be detected: nitrate reductase, β-galactosidase, β-glucosidase, esterase, esterase lipase, leucine arylamidase, cystine arylamidase, acid phosphatase, and phosphoamidase. Activities of pyrazinamidase, urease, pyrrolidonyl arylamidase, gelatinase, lipase, valine arylamidase, trypsin, chymotrypsin, α-galactosidase, β-glucuronidase, α-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase are not detected. The CAMP reaction is negative.
The cell wall contains meso-DAP. Mycolic acids are present. Cellular fatty acids are consistent with those described for the genus.
Source: a patient's wound after a dog bite.
DNA G+C content (mol%): not available.
Type strain: 1045, CCUG 56874, DSM 45254.
Sequence accession no. (16S rRNA gene): FJ157329.
Further comments: additional GenBank accession numbers include (partial rpoB gene) GQ913437.
Corynebacterium freneyi
(Renaud, Aubel, Riegel, Meugnier and Bollet 2001) emend. Funke and Frodl 2008a, 1515 (Effective publication: Funke and Frodl 2008a, 642.)
fre'ney.i. N.L. gen. masc. n. freneyi of Freney, to honor the French microbiologist Jean Freney.
Gram-stain-positive typically club-shaped rods. Colonies are whitish-grayish, but a minority of strains is yellowish, dry, rough, with a distinct wrinkled morphology and irregular edges. Colonies are 1–2 mm in diameter after 48 h of incubation on blood agar. Nonlipophilic. Fermentative metabolism. Reduction of nitrates is variable. The strains express α-glucosidase, pyrazinamidase, alkaline phosphatase, esterase, esterase lipase, leucine arylamidase (20 of 21 strains), and phosphoamidase. They do not produce pyrrolidonyl arylamidase, β-glucuronidase, β-galactosidase, N-acetyl-β-glucosaminidase, valine arylamidase, cystine arylamidase, trypsin, chymotrypsin, α-galactosidase, α-mannosidase, or α-fucosidase. The strains do not hydrolyze esculin, gelatin, or urea. The CAMP reaction is negative. Produces acid from glucose, maltose, sucrose, galactose (20 of 21 strains), D-fructose, D-mannose, trehalose, D-turanose, and 5-keto-gluconate. Ribose and D-tagatose acidifications are slow and weak. The results of fermentation of lactose and glycogen are variable, and very few strains (2 of 21) ferment mannitol. Xylose, glycerol, erythritol, D-arabinose, and 2-keto-gluconate are not fermented. Glucose is also fermented at 42°C, and strains may grow at 20°C.
The cell wall contains meso-DAP, sugars arabinose and galactose, and mycolic acids. Cellular fatty acids are consistent with those described for the genus.
Source: human clinical specimens.
DNA G+C content (mol%): not available.
Type strain: CCUG 45704, CIP 106767, DSM 44506, ISPB 6695110, JCM 12104.
Sequence accession no. (16S rRNA gene): AJ292762.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492237. Some strains are resistant to erythromycin, clindamycin, penicillin, and cefotaxime. Corynebacterium freneyi cannot be easily discerned from Corynebacterium xerosis and Corynebacterium hansenii by 16S rRNA, rpoB, or 16S–23S spacer region gene sequence analyses; these are best separated by DNA–DNA hybridization or phenotypic means.
Corynebacterium glaucum
Yassin, Kroppenstedt and Ludwig 2003, 707VP
glau'cum. L. neut. adj. glaucum bluish, light-gray, pertaining to the appearance of colonies.
Cells are Gram-stain-positive, dumbbell-shaped (when examined after 18 h growth), or showing typical coryneform morphology (when examined after 1 week of growth). On Columbia blood agar supplemented with 5% sheep blood, BHI agar and trypticase soy agar, colonies are light-gray in color. Facultatively anaerobic. Nitrate is not reduced. Hydrolyzes hippurate but not esculin, urea, gelatin, and starch. Produces acid from glucose and sucrose but not from arabinose, cellobiose, glycerol, glycogen, inulin, lactose, maltose, mannitol, raffinose, rhamnose, ribose, salicin, sorbitol, trehalose, and xylose. Displays alkaline phosphatase, pyrazinamidase, ester lipase, leucine arylamidase, and phosphoamidase activities but is negative for acid phosphatase, arginine dihydrolase, cystine arylamidase, esterase, lipase, α-glucosidase, β-glucosidase, α-galactosidase, β-galactosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, α-mannosidase, α-fucosidase, pyrrolidonyl arylamidase, valine arylamidase, trypsin, and chymotrypsin. Acetoin production is positive, but indole production is negative. Produces lactate but not propionate as the major product of glucose fermentation.
Contains meso-DAP in addition to sugars galactose and arabinose (i.e. cell-wall chemotype IV). Contains corynemycolic acids, and fatty acid profile contains saturated and unsaturated fatty acids. Tuberculostearic acid is absent. Contains MK-7(H2), MK-8(H2), and MK-9(H2) as respiratory menaquinones, with MK-8(H2) as the major component. Phospholipid pattern type PI, with no nitrogen-containing compounds.
Source: a cosmetic dye.
DNA G+C content (mol%): 64.3 (HPLC).
Type strain: IMMIB R-5091, DSM 44530, JCM 12208, NRRL B-24142.
Sequence accession no. (16S rRNA gene): AJ431634.
Corynebacterium glucuronolyticum
Funke, Bernard, Bucher, Pfyffer and Collins 1995b, 879VP (Effective publication: Funke, Bernard, Bucher, Pfyffer and Collins 1995a, 214.)
glu.cu.ro.no.ly'ti.cum. N.L. n. acidum glucuronicum glucuronic acid; N.L. neut. adj. lyticum (from Gr. neut. adj. lutikon) able to loosen, able to dissolve; N.L. neut. adj. glucuronolyticum cleaving β-glucuronic acid.
Gram-stain-positive coryneforms, about 1–3 µm in length. Colonies are white-yellowish, circular, convex, and nonhemolytic on sheep blood agar. Colonies are 1–1.5 mm after 24 h of incubation in 5% CO2 on blood agar. Nonlipophilic. Facultatively anaerobic. Acid is produced from glucose and sucrose; some strains produce acid from maltose, xylose, and ribose. Acid is not produced from mannitol and glycogen. Pyrazinamidase, esterase, esterase lipase, leucine arylamidase, cystine arylamidase, acid phosphatase, and, β-glucuronidase are present; some strains exhibit activity of alkaline phosphatase, phosphoamidase, α-glucosidase, and β-glucosidase. Hydrolysis of urea and esculin are variable. Some strains reduce nitrate.
Cellular fatty acids are consistent with those described for the genus.
Source: genitourinary specimens of human males, but can also be isolated from the urogenital tract of pigs (Devriese et al., 2000) or human body fluids that are normally sterile (Bernard et al., 2002).
DNA G+C content (mol%): 52–58 (HPLC).
Type strain: 6, ATCC 51860, CCUG 35055, CIP 104577, DMMZ 838, DSM 44120, JCM 11612, LMG 19047.
Sequence accession no. (16S rRNA gene): X86688.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492256. Corynebacterium glucuronolyticum strains may be resistant to erythromycin, clindamycin, and tetracycline (Funke et al., 1996b). Corynebacterium seminale (Riegel et al., 1996b) (Effective publication: Riegel et al. 1995d.) is deemed to be a later heterotypic synonym of Corynebacterium glucuronolyticum (Devriese et al., 2000). The DNA G+C content (mol%) of Corynebacterium seminale was found to be 60% (capillary electrophoresis method), but that of Corynebacterium glucuronolyticum sp. nov. was not extant in 1995. Corynebacterium seminale type strain has been curated under ATCC 51866, CCUG 34780, CCUG 34888, CIP 104297, DSM 44288, IBS B12915, JCM 10394, with GenBank accession number (16S rRNA gene) X84375.
Corynebacterium glutamicum
(Kinoshita, Nakayama and Akita 1958) Abe, Takayama and Kinoshita 1967, 299AL (Micrococcus glutamicus Kinoshita, Nakayama and Akita 1958, 176)
glu.ta'mi.c.um. N.L. neut. adj. glutamicum pertaining to glutamic acid.
(This description is largely based on that of Collins and Cummins, 1986.)
Short Gram-stain-positive rods or ellipsoids 0.7–1.0 × 1.0–3.0 µm, occurring singly, in pairs, or in irregular masses. In lag phase cultures, quite long branching cells, but in mid-to late exponential phase cultures the cells are very short, ellipsoidal to almost coccal. Colonies on nutrient agar are smooth, entire, circular, dull to slightly glistening, and generally pale yellow to yellow. Gray to black colonies on tellurite agar. Moderate turbidity with floccular sediment in broth. Optimal growth at 25–37°C, with only slight growth at 42°C. Aerobic and facultatively anaerobic. Glucose, fructose, mannose, maltose, sucrose, trehalose, and methyl red positive. Negative for arabinose, xylose, rhamnose, galactose, lactose, raffinose, salicin, dextrin, and starch fermentation. Hippurate hydrolyzed, urease is produced, but gelatin is negative. Nitrate is reduced but phosphatase and pyrazinamidase negative. Esculin and casein negative.
Cell wall contains meso-DAP and the sugars arabinose and galactose. Mycolates are present. MK-9(H2) is the major menaquinone. Cellular fatty acids consistent with those of the genus. All strains produce large amounts of L-glutamic acid under aerobic conditions.
Source: sewage.
DNA G+C content (mol%): 55–57.7 (Tm), 53.8 (complete genome sequencing) (Ikeda and Nakagawa, 2003; Kalinowski et al., 2003).
Type strain: ATCC 13032, CCUG 27702, CIP 82.8, DSM 20300, HAMBI 2052, JCM 1318, LMG 3730, NBRC 12168, NRRL B-2784.
Sequence accession no. (16S rRNA gene): AF314192.
Further comments: additional GenBank accession numbers include (complete rpoB gene) NC_003450. Brevibacterium divaricatum Su and Yamada 1960AL, Corynebacterium lilium Lee and Good 1963, 1349AL, “Brevibacterium flavum” DSM 20411, and “Brevibacterium lactofermentum” DSM 20412 and DSM 1412 are all considered as later heterotypic synonyms of Corynebacterium glutamicum (Liebl et al., 1991). Complete genome sequence of Corynebacterium glutamicum ATCC 13032T was deposited under GenBank accession no. NC_006958 with 3,282,708 bp, from which various transcriptional functions were conjectured (Brune et al., 2005; Kalinowski et al., 2003). Concurrently, a second genome sequencing project of the same strain, deposited under GenBank accession no. NC_004350, was described as having 3,309,401 bp (Ikeda and Nakagawa, 2003).
Corynebacterium halotolerans
Chen, Li, Tang, Kroppenstedt, Stackebrandt, Xu and Jiang 2004, 781VP
ha.lo.to'le.rans. Gr. n. hals halos salt; L. part. adj. tolerans tolerating; N.L. pres. part. halotolerans referring to the ability to tolerate high salt concentrations.
Gram-stain-positive diphtheroid and irregular rods. Colonies on modified ISP 5 medium, trypticase/soy agar medium, and Mueller–Hinton agar medium are moderately yellow, circular, entire, somewhat convex, and opaque after 24 h at 28°C. Optimum growth temperature is 28°C. Obligately aerobic. Nonlipophilic. Optimum gowth concentration of KCl, NaCl, and MgCl2·6H2O is 10%. Positive for nitrate reduction, but negative for milk peptonization and coagulation, gelatin liquefaction, growth in cellulose, production of H2S and melanin, starch hydrolysis, and urease production. Activities for lipase and β-glucuronidase are positive. Ornithine decarboxylase, arginine dihydrolase, lysine decarboxylase, α- and β-galactosidase, N-acetyl-β-glucosaminidase, and β-glucosidase activities are negative. The following substrates are utilized: glucose, galactose, sucrose, arabinose, mannose, mannitol, maltose, starch, xylose, ribose, salicin, and dextrin. Cellobiose, fructose, amygdalin, and lactose are not utilized. Acid production occurs only from glucose.
The cell wall contains meso-DAP with sugars mainly galactose and arabinose. Menaquinones are MK-8(H2) (35.5%) and MK-9(H2) (64.5%); phospholipids are diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol, glycolipid, and phosphatidylinositol mannosides. Cellular fatty acids are consistent with those described for the genus, with significant quantities (7.4% of total) TBSA detected. Predominant mycolic acids include C32:0 (36.0%), C34:0 (20.8%), C34:1 (25.1%), C36:0 (3.6%), C36:1 (8.4%), and C36:2 (5.1%).
Source: saline soil collected in Xinjiang Province, west China.
DNA G+C content (mol%): 63 (Tm).
Type strain: YIM 70093, CCTCC AA 001024, DSM 44683, JCM 12676.
Sequence accession no. (16S rRNA gene): AY226509.
Further comments: additional GenBank accession numbers include (partial rpoB gene) EU004065.
Corynebacterium hansenii
Renaud, Le Coustumier, Wilhem, Aubel, Riegel, Bollet and Freney 2007, 1115VP
han.sen'i.i. N.L. gen. masc. n. hansenii of Hansen, to honor the Belgian microbiologist Willy Hansen.
Gram-stain-positive, typically club-shaped rods. Colonies are yellow-pigmented, dry, rough, and with irregular edges, roughly 0.5–1.0 mm in diameter. Nonlipophilic. Facultatively anaerobic. Produces acid from glucose, ribose, maltose, and sucrose. Does not reduce nitrates. No hydrolysis of esculin, urea, or gelatin. Pyrazinamidase positive. Does not produce alkaline phsophatase, pyrrolidonyl arylamidase, β-glucuronidase, β-galactosidase, α-glucosidase, or N-acetyl-β-glucosaminidase. D-Ribose, D-galactose, D-glucose, and D-fructose are used as carbon substrates, but D-maltose and D-turanose are not. Glucose is not fermented at 42°C but growth will occur at 20°C.
Source: human liposarcoma pus; the pathogenic role is not known.
DNA G+C content (mol%): not available.
Type strain: C-138, CCUG 53252, CIP 108444, DSM 45109, JCM 15293.
Sequence accession no. (16S rRNA gene): AM946639.
Further comments: additional GenBank accession numbers include (partial rpoB gene) AY684044 and FJ268580. Corynebacterium hansenii cannot be easily discerned from Corynebacterium xerosis and Corynebacterium freneyi by 16S rRNA, rpoB, or 16S–23S spacer region gene sequence analyses; these are best separated by DNA–DNA hybridization or phenotypic means.
Corynebacterium imitans
Funke, Efstratiou, Kuklinska, Hutson, De Zoysa, Engler and Collins, 1997a, 1274 (Effective publication: Funke, Efstratiou, Kuklinska, Hutson, De Zoysa, Engler and Collins 1997b, 1982.)
i'mi.tans. L. part. adj. imitans imitating, copying, indicating that this bacterium was isolated from patients with an illness that was imitating the clinical picture of pharyngeal diphtheria and also indicating that the biochemical profile of the newly described bacterium imitates the biochemical profiles of other Corynebacterium species.
Gram-stain-positive diphtheroids. Nonlipophilic. Colonies are white-grayish, glistening, circular, convex, creamy, and have entire edges. Acid is produced from D-arabinose, ribose, D-glucose, D-fructose, D-mannose, maltose, lactose, and L-fucose and weakly from sucrose but not from glycerol, erythritol, L-arabinose, xylose, adonitol, β-methyl-xyloside, galactose, L-sorbose, rhamnose, dulcitol, inositol, mannitol, sorbitol, α-methyl-mannoside, α-methyl-glucoside, N-acetylglucosamine, amygdalin, arbutin, salicin, cellobiose, melibiose, trehalose, inulin, melezitose, D-raffinose, glycogen, xylitol, b-gentiobiose, D-turanose, D-lyxose, D-tagatose, D-fucose, D-arabitol, L-arabitol, gluconate, and 2-keto-gluconate. Nitrate reduction is variable (Funke, unpublished). Urea, esculin, and tyrosine are not hydrolyzed. The CAMP reaction is positive. Pyrazinamidase (weak), alkaline phosphatase, esterase, esterase lipase, acid phosphatase, and phosphoamidase activities are detected, but lipase, pyrrolidonyl arylamidase, leucine arylamidase, valine arylamidase, trypsin, chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase activities are not detected.
Meso-DAP and mycolic acids are present. Cellular fatty acids are consistent with those described for the genus. Does not produce propionic acid as metabolic product (Bernard et al., 2002).
Source: human clinical specimens.
DNA G+C content (mol%): 62 (Tm).
Type strain: ATCC 700354, CCUG 36877, CIP 105130, DSM 44264, IFO (now NBRC) 16163, JCM 10386, NBRC 100416, NCTC 13015.
Sequence accession no. (16S rRNA gene): Y09044.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492259.
Corynebacterium jeikeium
Jackman, Pitcher, Pelczynska and Borman 1988, 136VP (Effective publication: Jackman, Pitcher, Pelczynska and Borman 1987, 88.)
jei.kei'um. N.L. neut. adj. jeikeium formed from the initial letters of the surnames of W. D. Johnson and D. Kaye, doctors who described infections attributed to the taxon in 1970.
Gram-stain-positive rods, ∼0.5 × 2 µm, pleomorphic, occasionally club-shaped, arranged in V-forms or palisades. Cells contain metachromatic granules. Colonies are small (1–2 mm), entire, grayish-white, and nonhemolytic. Obligate aerobic. Lipophilic. Good growth occurs at 30–42°C but poorly at 22°C. Grows on 0.03% tellurite agar and bile salt agar, hydrolyzes Tweens 20 and 80 but not Tweens 40 and 60. Acid is produced from glucose and galactose; some strains produce acid from maltose. Acid is not produced from adonitol, arabinose, cellobiose, dulcitol, dextrin, erythritol, fructose, glycerol, glycogen, inositol, lactose, mannitol, mannose, melibiose, raffinose, rhamnose, salicin, sorbose, sucrose, and trehalose. Methyl red test negative; nitrate is not reduced. H2S, acetoin, and indole are not produced. Tyrosine, casein, gelatin, and urea are not degraded. Citrate is not utilized. Esculin and starch are not hydrolyzed, gluconate is not converted to 2-ketogluconate. Arginine dihydrolase, and lysine and ornithine decarboxylase tests are negative.
Cell wall contains meso-DAP and sugars arabinose and galactose. Mycolic acids are present and are principally of carbon-chain length C32–C36. Cellular fatty acids are consistent with those described for the genus with only trace amounts of 10-methyl-octadecanoic acid (tuberculostearic acid) occurring. Does not produce propionic acid as metabolic product (Bernard et al., 2002).
Source: human body surfaces and human clinical specimens.
DNA G+C content (mol%): 58–61 (Tm).
Type strain: ATCC 43734, CCUG 27192, CIP 103337, DSM 7171, JCM 9384, NCTC 11913.
Sequence accession no. (16S rRNA gene): X84250.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492231.
Corynebacterium jeikeium Jackman et al. 1988 was previously known as CDC coryneform group JK. Four genomo-species among Corynebacterium jeikeium strains have been described (Riegel et al., 1994b). Corynebacterium jeikeium strains are often multidrug-resistant (Riegel et al., 1996a; Sanchez et al., 2003). The complete genome of Corynebacterium jeikeium strain K411 deposited under GenBank accession no. CR931997 has 2,462,499 bp (Tauch et al., 2005).
Corynebacterium kroppenstedtii
Collins, Falsen, Åkervall, Sjödén and Alvarez 1998, 1453VP
krop.pen.sted'ti.i. N.L. gen. masc. n. kroppenstedtii of Kroppenstedt, to honor the German microbiologist Reiner M. Kroppenstedt for his many contributions to the microbiology of actinomycetes.
Gram-stain-positive diphtheroids that occur as single cells or are arranged in V-shaped forms or palisades. Colonies on blood agar are nonpigmented, small (<0.5 in diameter after 24 h incubation), smooth, convex, and nonhemolytic. Facultatively anaerobic. Grows in 10% NaCl and at 42°C. Nitrate is not reduced to nitrite. Esculin is hydrolyzed but urea and gelatin are not. Acid is produced from glucose, maltose (weak reaction), and sucrose. Acid is not produced from lactose, ribose, D-xylose, mannitol, and glycogen. Leucine arylamidase, esterase, esterase lipase, and pyrazinamidase are produced. Alkaline and acid phosphatases, N-acetyl-β-glucosaminidase, cystine arylamidase, chymotrypsin, α-fucosidase, α-galactosidase, β-galactosidase, β-glucosidase, β-glucuronidase, lipase, α-mannosidase, pyrrolidonyl arylamidase, trypsin, and valine arylamidase are not detected.
Cell wall contains meso-DAP and the sugars arabinose and galactose Cellular fatty acids are consistent with those described for the genus, with tuberculostearic acid being detected. Mycolic acids are not present. Some clinical isolates produce propionic acid as a metabolic product (Bernard et al., 2002).
Source: human clinical specimens, in particular, in cases of lobular granulomatous mastitis (Kieffer et al., 2006; Paviour et al., 2002; Riegel et al., 2004).
DNA G+C content (mol%): 62 (Tm).
Type strain: CCUG 35717, CIP 105744, DSM 44385, JCM 11950.
Sequence accession no. (16S rRNA gene): Y10077.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492274. Although Corynebacterium kroppenstedtii was not described as being lipophilic by Collins et al., this feature has been observed by other authors (Riegel et al., 2004; Tauch et al., 2008a). The complete genome of DSM 44385T, with 2,446,804 bp, has been deposited under GenBank accession no. NC_012704 (Tauch et al., 2008a).
Corynebacterium kutscheri
(Migula 1900) Bergey, Harrison, Breed, Hammer and Huntoon 1925, 395AL (Bacterium kutscheri Migula 1900, 372)
kut'sche.ri. N.L. gen. masc. n. kutscheri of Kutscher, the bacteriologist who first isolated the species.
(This description is largely based on that of Collins and Cummins, 1986.)
Irregularly staining slender rods, often clubbed, sometimes with pointed ends; metachromatic granules present. Large numbers of fimbriae present. Small, thin, yellowish or grayish white serrate colonies on nutrient agar. Facultatively anaerobic. Reduces potassium tellurite. Positive for glucose, fructose, mannose, maltose, sucrose, salicin, dextrin, and starch; negative for arabinose, xylose, rhamnose, galactose, lactose, and raffinose. Trehalose variable. Nitrate is reduced, urease is produced, hippurate is hydrolyzed and pyrazinamidase is detected. Tyrosine, esculin, and gelatin are not hydrolyzed; casein is not digested; methyl red and phosphatase are negative.
Cell wall contains meso-DAP and the sugars arabinose, galactose, mannose, and rhamnose. This species appears to be the only organism in the genus Corynebacterium which may have rhamnose as a cell-wall sugar in addition to arabinose and galactose. Metabolic products include propionate and lactate, with lesser amounts of acetate, pyruvate, and oxalacetate.
Source: a frequent parasite of mice and rats, but also occurs in other small rodents such as voles. An infection in a human has been reported after the patient had been bitten by a rat (Holmes and Korman, 2007).
DNA G+C content (mol%): ∼46 (Tm).
Type strain: ATCC 15677, CCUG 27535, CIP 103423, DSM 20755, JCM 9385, NBRC 15288, NCTC 11138.
Sequence accession no. (16S rRNA gene): X81871.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492257.
Corynebacterium lipophiloflavum
Funke, Hutson, Hilleringmann, Heizmann and Collins 1997a, 1274VP (Effective publication: Funke, Hutson, Hilleringmann, Heizmann and Collins 1997c, 223.)
li.po.phi.lo.fla'vum. Gr. n. lipos fat; Gr. neut. adj. philon loving; L. adj. flavus -a -um yellow; N.L. neut. adj. lipophiloflavum fat loving and yellow.
Gram-stain-positive club-shaped rods, 1–3 µm in length. Slightly lipophilic. Colonies are yellowish, circular, convex, and slightly dry; tiny colonies (0.2 mm in diameter) after 24 h incubation on blood agar, but ∼1 mm if grown on blood agar enriched with 1% Tween 80. Acid is not produced from glucose, maltose, sucrose, mannitol, xylose, lactose, and glycogen. Nitrate is not reduced. Urea is slowly hydrolyzed, but esculin is not hydrolyzed. The CAMP reaction is negative. Pyrazinamidase, alkaline phosphatase, esterase, esterase lipase, lipase, leucine arylamidase, acid phosphatase, and phosphoamidase activities are detected but pyrrolidonyl arylamidase, valine arylamidase, cystine arylamidase, trypsin, chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase activities are not detected.
Cell wall contains meso-DAP. Mycolic acids are present. Cellular fatty acids are consistent with those described for the genus.
Source: human clinical specimens.
DNA G+C content (mol%): 65 (Tm).
Type strain: ATCC 700352, CCUG 37336, CIP 105127, DMMZ 1944, DSM 44291, JCM 10383.
Sequence accession no. (16S rRNA gene): Y09045.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492260.
Corynebacterium lubricantis
Kämpfer, Lodders, Warfolomeow, Falsen and Busse 2009, 1114VP
lu.bri.can'tis. L. v. lubricare to lubricate; N.L. lubricans – antis (from L. part. adj. lubricans); N.L. gen. n. lubricantis of/from a (coolant) lubricant.
Gram-stain-positive rods ∼1 µm long, with yellow, shiny, translucent colonies 0.5 mm in diameter after 24 h. Grows from 10–45°C but most testing was done at 28°C. Unique, as this species can grow on MacConkey agar. The following compounds are used as sole sources of carbon: D-fructose, D-glucose, acetate, fumarate, and DL-lactate; the following compounds are not utilized: N-acetylgalactosamine, N-acetylglucosamine, L-arabinose, L-arbutin, cellobiose, D-galactose, maltose, D-mannose, L-rhamnose, D-ribose, salicin, trehalose, D-xylose, adipate, 2-oxoglutarate, D-gluconate, melibiose, sucrose, adonitol, myo-inositol, maltitol, D-mannitol, D-sorbitol, propionate, cis- and trans-aconitate, 4-aminobutyrate, citrate, glutarate, DL-3-hydroxybutyrate, itaconate, L-malate, mesaconate, pyruvate, L-alanine, D-alanine, L-aspartate, L-leucine, L-ornithine, L-proline, L-serine, putrescine, azelate, suberate, L-histidine, L-phenylalanine, L-serine, L-tryptophan, 3-hydroxybenzoate, and phenylacetate. Acid is produced from glucose and trehalose. No acids are produced from sucrose, D-mannitol, dulcitol, lactose, rhamnose, maltose, galactose, salicin, adonitol, inositol, sorbitol, L-arabinose, raffinose, D-xylose, trehalose, cellobiose, methyl-D-glucoside, erythritol, melibiose, D-arabitol, or D-mannose. p-Nitrophenyl (pNP)-β-D-glucopyranoside, p-NP-β-D-glucuronide, bis-p-NP-phosphate, and bis-p-NP-phenylphosphonate are hydrolyzed. The following compounds are not hydrolyzed: p-NP-α-D-glucopyranoside, p-NP-β-D-galactopyranoside, p-NP-β-D-xylopyranoside, bis-p-NP-phosphorylcholine, L-alanine-p-nitroanilide (pNA), L-proline pNA, and C-L-glutamate pNA.
Chemotaxonomic properties as described for genus except that in addition to MK-9(H2) and MK-8(H2), MK-7(H2), is also detected. The lipid profile consists of the major compounds phosphatidylglycerol and an unknown glycolipid, moderate amounts of phosphatidylinositol, diphosphatidylglycerol, two unknown glycolipids, three unknown aminolipids and an unknown polar lipid, and minor amounts of two glycolipids, an aminolipid and a polar lipid. Polyamines are present in small amounts; the major compounds are spermidine and spermine. The characteristic peptidoglycan diamino acid is meso-DAP. Testing for corynemycolates was not done. Cellular fatty acids are consistent with those described for the genus except that significant (11–31% of total vol.) of tuberculostearic acid is detected.
Source: coolant lubricant.
DNA G+C content (mol%): 66.6 (HPLC).
Type strain: KSS-3Se, CCM 7546, CCUG 56567, DSM 45231, JCM 16607.
Sequence accession no. (16S rRNA gene): FM173119.
Corynebacterium macginleyi
Riegel, Ruimy, De Briel, Prévost, Jehl, Christen and Monteil 1995e, 132VP
mac.gin'ley.i. N.L. gen. masc. n. macginleyi of McGinley, named in honor of Kenneth John McGinley who made important contributions to the knowledge of lipid-requiring diphtheroids.
Gram-stain-positive pleomorphic rods occasionally with metachromatic granules, arranged in palisades or V-shaped forms. Facultatively anaerobic. Strains are lipophilic in that no growth is seen in brain heart infusion broth after 72 h at 37°C, but growth is visible after 24 h in the same broth supplemented with 0.01–1% Tween 80. Very small (less than 1 mm in diameter) and non-β-hemolytic colonies appear after 48 h on sheep blood agar, but on 1% Tween 80-supplemented sheep blood agar reddish-beige colonies are larger (2–4 mm in diameter) and Tween esterase activity is noted. Nitrate is reduced to nitrite. Acid is produced from D-glucose, ribose, and sucrose but not from maltose, lactose, D-xylose, trehalose, or glycogen. Fermentation of D-mannitol occurs variably. Tyrosine, gelatin, starch, and urea are not degraded or hydrolyzed. Esculin is not hydrolyzed. DNA is degraded. Hydrolysis of hippurate is variable. Arginine is not degraded. Cells produce alkaline phosphatase, but pyrazinamidase, pyrrolidonyl, arylamidase, β-glucuronidase, β-galactosidase, α-glucosidase, and N-acetyl-β-glucosaminidase are not produced.
Cell wall contains meso-DAP and the sugars arabinose and galactose. Mycolic acids of short chain lengths (C26–C36) are present cellular fatty acid composition consistent with those of the genus, but tuberculostearic acid was not detected. Propionic acid is not detected as metabolic product (Bernard et al., 2002).
Source: predominately recovered from human eye specimens, particularly the conjunctiva (Funke et al., 1998d; Joussen et al., 2000) and from a case of endophthalmitis (Ferrer et al., 2004).
DNA G+C content (mol%): 58 (HPLC).
Type strain: JCL-2, ATCC 51787, CCUG 32361, CIP 104099, DSM 44284, JCM 11684.
Sequence accession no. (16S rRNA gene): AJ439345, X80499.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492276. Corynebacterium macginleyi Riegel et al. 1995d was previously known as CDC coryneform group G-1, the nitrate reducing form of CDC coryneform group G as well as a lipid-requiring diphtheroid, genomospecies II. Opportunistic infections in humans by Corynebacterium macginleyi associated with other sites have been described, including intravenous catheter-related infections (Dobler and Braveny, 2003; Villanueva et al., 2002) and endocarditis (Pubill Sucarrat et al., 2003). An outbreak involving quinolone-resistant strains has been reported (Eguchi et al., 2008). rpoB Gene sequencing should be done to differentiate strains from Corynebacterium accolens, as these species can not be easily distinguished by 16S rRNA gene sequencing.
Corynebacterium marinum
Du, Jordan, Rooney, Chen and Austin 2010, 1946VP
ma.ri'num. L. neut. adj. marinum of the sea, marine.
Cells are short Gram-stain-positive diphtheroid rods; some of the cells are arranged in a V formation. Colonies on Marine 2216E agar medium are circular, erose, convex, yellow, of a creamy consistency, and ∼0.5–1.5 mm after 48 h at 28°C. Facultatively anaerobic; methyl red negative. Voges–Proskauer positive, nitrate is reduced, and horse blood cells are lyzed. Esculin and urea are not hydrolyzed, but casein is digested, and starch and pullulan are hydrolyzed. Gelatin is not liquefied. Tween 20–80 are hydrolyzed. Cells grow in the presence of 0–8% (w/v) NaCl and at 4–37°C. Prolific growth occurs at 30–32°C in media that contains 1% (w/v) NaCl. Using the API 50CH system, esculin ferric, citrate, salicin, D-maltose, and glycogen are utilized. Acid is produced from glucose, maltose, sucrose, and glycogen, but not from ribose, xylose, mannitol, or lactose. Esterase lipase, leucine arylamidase, α-chymotrypsin, naphthol-AS-BI-phosphohydrolase, pyrazinamidase, and β-glucuronidase are positive. Esterase, lipase, valine arylamidase, cystine arylamidase, trypsin, alkaline phosphatase, pyrrolidonyl arylamidase, acid phosphatase, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase are negative. The antibiotic susceptibility of the isolate was tested using antimicrobial discs. The type strain is resistant to nalidixic acid (30 µg), nitrofurantoin (50 µg), sulfamethizole (200 µg), tetracycline (100 µg), and cotrimoxazole (25 µg), but sensitive to ampicillin (25 µg), chloramphenicol (50 µg), gentamicin (10 µg), kanamycin (30 µg), carbenicillin 143 (100 µg), and streptomycin (25 µg).
Cellular fatty acid composition is consistent with those of the genus. Detection of corynemycolates and other chemotaxonomic information not described.
Source: coastal sediment close to a coal-fired power station in Qingdao, China.
DNA G+C content (mol%): 65.0 (HPLC).
Type strain: D7015, CGMCC 1.6998, NRRL B-24779.
Sequence accession no. (16S rRNA gene): DQ219354.
Corynebacterium maris
Ben-Dov, Ben Yosef, Pavlov and Kushmaro 2009, 2461VP
ma'ris. L. gen. n. maris of the sea.
Gram-stain-positive coccobacilli approximately 0.5–0.8 µm × 0.8–1.5 µm. Nonhemolytic. Forms small colonies (approximately 1 mm and 2.5 mm in diameter after 48 and 72 h, respectively) after incubation at 30°C. Colonies are yellowish to yellow, circular, convex, smooth, and opaque. Grows well at 0.5–4.0% salinity, at pH 7.2–9.0, and at 30–37°C. Aerobic. Oxidase positive. Alkaline phosphatase, esterase, esterase lipase, lipase, leucine arylamidase, α-glucosidase, pyrazinamidase, pyrrolidonyl arylamidase, and gelatin hydrolysis detected. No activity is observed for nitrate reduction, valine and cystine arylamidases, trypsin, α-chymotrypsin, acid phosphatase, naphthol-AS-BI-phosphohydrolase, α- and β-galactosidases, β-glucuronidase, β-glucosidase, urease, N-acetyl-β-glucosaminidase, α-mannosidase, or α-fucosidase. The strain oxidizes the following carbon compounds as sole energy sources: maltose, lactulose, β-hydroxybutyric acid, α-ketovaleric acid, Tween 40, phenylethylamine, N-acetyl-D-galactosamine, malonic acid, L-threonine, L-glutamic acid, L-fucose, L-alanyl glycine, inosine and, less efficiently, raffinose, D-arabitol, L-asparagine, and citric acid, as determined with the Biolog GN system.
Cellular fatty acids are consistent with those described for the genus, with tuberculostearic acid detected. Mycolic acids (C30–C36) are present.
Source: the mucus of the coral Fungia granulosa, Gulf of Eilat, Red Sea.
DNA G+C content (mol%): 66.6 (HPLC).
Type strain: Coryn-1, DSM 45190, JCM 17108, LMG 24561.
Sequence accession no. (16S rRNA gene): FJ423600.
Corynebacterium massiliense
Merhej, Falsen, Raoult and Roux 2009, 1958VP
mas.si.li.en'se. L. neut. adj. massiliense of or pertaining to Massilia, the old Roman name for Marseille from where the type strain was isolated.
Gram-stain-positive club-shaped rods, occurring as single cells, in pairs, or small clusters, 0.2–1.8 µm in length and 0.3–0.7 µm in diameter after 48 h growth in TSB medium. Colonies are circular, grayish, glistening, and 0.5–1 mm in diameter after growth on blood agar for 48 h. Capable of aerobic growth with only weak growth anaerobically and microaerophilically. Temperature range 30–44°C (optimum, 37°C). With the API Coryne system, acid is not produced from D-glucose, D-ribose, D-xylose, D-mannitol, maltose, D-lactose, sucrose, or glycogen. Nitrate is not reduced. Urea, gelatin, and esculin are not hydrolyzed. With API Coryne and API ZYM strips, positive for leucine arylamidase, weakly positive for pyrazinamidase, alkaline phosphatase, esterase and esterase lipase, but negative for pyrrolidonyl arylamidase, lipase, valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, acid phosphatase, naphthol-AS-BI-phosphohydrolase, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase.
Cellular fatty acids are consistent with those described for the genus, and tuberculostearic acid may be detected. Testing for corynemycolates or other cell wall markers not done.
Source: human clinical materials.
DNA G+C content (mol%): not available.
Type strain: 5402485, CCUG 53857, CIP 109423, CSUR P19.
Sequence accession no. (16S rRNA gene): EF217056.
Further comments: additional GenBank accession numbers include (partial rpoB gene) EF217057.
Corynebacterium mastitidis
Fernández-Garayzábal, Collins, Hutson, Fernandez, Monasterio, Marco and Dominguez 1997, 1084VP
mas.ti'ti.dis. N.L. gen. n. mastitidis of an inflammation of the mammary gland.
Gram-stain-positive rods occurring singly, in palisades, or V-shaped forms. On blood agar, small (diameter less than 1 mm), rough, whitish, low convex colonies formed after 3 d of incubation at 37°C. Nonhemolytic. Esculin and gelatin not hydrolyzed, and nitrate not reduced. Hydrolysis of urea variable. Acid is not produced from glucose, ribose, xylose, mannitol, lactose, maltose, sucrose, and glycogen. Pyrazinamidase, alkaline phosphatase, acid phosphatase, esterase, esterase lipase, lipase, leucine arylamidase, valine arylamidase, and cystine arylamidase are produced. Pyrrolidonylarylamidase, β-glucuronidase, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, α-mannosidase, α-fucosidase, N-acetyl-p-glucosaminidase, trypsin, and chymotrypsin are not produced. The strains do not grow in the presence of 6.5% NaCl. The type strain has characteristics of the species described above except that it is urease and naphthol-AS-BI-phosphate positive.
The cell wall contains meso-DAP. Short-chain mycolic acids are present. The major long-chain fatty acids are C16:0, C18:0, and C18:1 ω9c. Tuberculostearic acid is not produced.
Source: milk and udder of infected sheep, but strains genetically most like Corynebacterium mastitidis have been found to cause human ocular infections (Eguchi et al., 2008).
DNA G+C content (mol%): not available.
Type strain: S-8, CCUG 38654, CECT 4843, CIP 105509, DSM 44356, JCM 12269, LMG 19040, NBRC 16160.
Sequence accession no. (16S rRNA gene): Y09806.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492281. Sheep strains are susceptible to penicillin G, ampicillin, amoxycillinclavulanic acid, gentamicin, cephalothin, and nalidixic acid. However, the strains described by Eguchi, which by 16S rRNA gene sequencing were closest to (98.2% identity) Corynebacterium mastitidis from human ocular infections, were generally susceptible to antimicrobics tested (Eguchi et al., 2008).
Corynebacterium matruchotii
(Mendel 1919) Collins 1983, 438VP (Effective publication: Collins 1982a, 365.) (Cladothrix matruchoti Mendel 1919, 584; Bacterionema matruchotii Gilmour, Howell and Bibby 1961, 139)
ma.tru.cho'ti.i. N.L. gen. masc. n. matruchotii of Matruchot; named after Professor Matruchot, a French mycologist.
(This description also includes information from Collins and Cummins, 1986.)
Cells are pleomorphic, comprising nonseptate and septate filaments and bacilli. Characteristic morphology is a bacillus attached to a filament (“whip-handle”). Branching is frequent with aerobic and/or acidic conditions. Metachromatic granules formed. Colony appearance is variable; aerobically incubated colonies are 0.5–1.5 mm in diameter, circular, convex, rough with entire or filamentous margin or irregular “molar-toothed” rough colonies. May be 1–2 mm and adherent if grown anaerobically. Corynebacterium matruchotii colonies at 37°C on TBSA have also been described as being 1 mm in diameter, white, creamy or gray, with smooth matte surface and creamy texture; some strains have a crinkled surface and can be lifted as a single dry colony when touched by a loop (Rassoulian-Barrett et al., 2001). Facultatively anaerobic. Optimal temperature 37°C. Complex nutritional requirements. Positive for glucose, fructose, mannose, maltose, sucrose, salicin, and dextrin. Negative for arabinose, xylose, rhamnose, galactose, lactose, and trehalose. Raffinose and starch fermentation variable. Nitrate is reduced and hippurate hydrolyzed. Gelatin is hydrolyzed, and casein is not digested; methyl red, and phosphatase are negative. Esculin and urea hydrolysis described originally as being positive or occasionally positive. Rassoulian-Barrett et al., based on study of genetically characterized strains, found that Corynebacterium matruchotii isolates were conventional tube esculin test negative but could be esculin positive when using the API CORYNE strip; strains were urease and mannitol negative by both methods (Rassoulian-Barrett et al., 2001). The production of pyrrolidonyl arylamidase was the only trait that consistently distinguished Corynebacterium matruchotii strains from Corynebacterium durum strains which are otherwise very similar to each other biochemically (Rassoulian-Barrett et al., 2001).
meso-DAP is detected, and arabinose and galactose are cell-wall sugars. Cellular fatty acids were consistent with those described for the genus, but tuberculostearic acid is not detected. Species contains corynemycolates. Principal menaquinones are MK-9(H2) and MK-8(H2). Polar lipids include diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol, phosphatidylinositol dimannosides, and unidentified glycolipids. Propionic acid is produced as a metabolic product (Riegel et al., 1997a).
Source: the oral cavity, particularly from calculus and plaque deposits; is otherwise an occasional pathogen of humans or primates.
DNA G+C content (mol%): 55–58 (Tm).
Type strain: ATCC 14266, CCUG 27545, CCUG 46620, CIP 81.82, DSM 20635, JCM 9386, NBRC 15360, NCTC 10254.
Sequence accession no. (16S rRNA gene): X82065, X84443.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492238.
Corynebacterium minutissimum
(ex Sarkany, Taplin and Blank 1962) Collins and Jones 1983a, 870VP emend. Yassin, Steiner and Ludwig 2002a, 1004
mi.nu.tis'si.mum. L. sup. neut. adj. minutissimum very small.
Colonies on blood agar are circular, about 1 mm in diameter after 24 h, circular, slightly convex, shiny, moist, and not pigmented. When grown in the presence of rich media (e.g. 20% fetal calf serum), colonies may demonstrate a coral-red to orange fluorescence when illuminated by a Wood's lamp (365 nm). A coralloid precipitin in agar is formed after growth on BHI agar supplemented with 1% Tween 80 whether in air or in CO2. No hemolysis on blood agar. Cells described as short, straight, or slightly curved rods (1–2 × 0.3–0.6 µm) arranged at right angles to give V-formations. Metachromatic granules may be observed. Facultatively anaerobic. Optimal temperature 37°C. Positive for glucose, fructose, maltose, mannose, hippurate, alkaline phosphatase, pyrazinamidase, and leucine arylamidase. Negative for xylose, lactose, raffinose, trehalose, starch, adonitol, amygdalin, arabinose, cellobiose, glycerol, glycogen, inulin, mannitol, melezitose, rhamnose, ribose, salicin, and sorbitol. Hippurate, sucrose, and mannose described as positive or variable but later as negative, positive, and negative, respectively. Acetoin positive, but indole negative; nitrate is not reduced, and urease is not produced. Tyrosine hydrolyzed (Funke and Bernard, 2007), but esculin and gelatin are not. Casein is not digested and methyl red is negative. Acid phosphatase, arginine dihydrolase, esterase, ester lipase, lipase, α-glucosidase, β-glucosidase, α-galactosidase, β-galactosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, α-mannosidase, α-fucosidase, naphthol-AS-BL-phosphoydrolase, pyrrolidonyl arylamidase, valine arylamidase, cystine arylamidase, trypsin, and chymotrypsin were negative.
Cells contain meso-DAP with arabinose and galactose as cell-wall sugars. Menaquinones MK-8(H2) and MK-9(H2) are detected. Diphosphatidylglycerol, phosphatidylinositol, phosphatidylinositol mannoside, and unidentified glycolipids are found. Corynemycolates are present (De Briel et al., 1992). Cellular fatty acids are consistent with those of the genus, and tuberculostearic acid is detected (Bernard et al., 1991). Lactic, but not propionic, acid is produced as metabolic product.
Source: a rare opportunistic human pathogen.
DNA G+C content (mol%): 56–58 (HPLC).
Type strain: ATCC 23348, CCUG 541, CIP 100652, DSM 20651, JCM 9387, NBRC 15361, NCTC 10288.
Sequence accession no. (16S rRNA gene): X82064, X84678, X84679.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492235. Accession number for NCTC 10288 X84678 was described by the depositor as Corynebacterium minutissimum clone 1 and NCTC 10288 X84679 as clone 2, respectively. By 16S rRNA gene sequencing, Corynebacterium minutissimum is closely related to (>98.9–99% identity) Corynebacterium aurimucosum and Corynebacterium singulare (Riegel et al., 1997b), but differs from these (∼93.8–94.6% identity, respectively) when partial or complete sequences of the rpoB gene are compared (Khamis et al., 2004). Corynebacterium minutissimum had long been attributed as a cause of erythrasma, a skin infection, but this inference has been challenged, as most publications citing this association did so in the absence of laboratory-based evidence (Coyle and Lipsky, 1990). It has been thought that some published Corynebacterium minutissimum reports in the past may actually represent misidentified Corynebacterium amycolatum strains (Zinkernagel et al., 1996).
Corynebacterium mucifaciens
Funke, Lawson and Collins 1997d, 956VP
mu.ci.fa'ci.ens. L. n. mucus slime; L. part. adj. faciens producing; N.L. part. adj. mucifaciens slime producing.
Colonies on blood agar are highly unusual with respect to members of this genus, as they are circular, convex, glistening, yellowish, uniquely mucoid, and about 1–1.5 mm in diameter after 24 h incubation at 37°C in 5% CO2. Facultatively anaerobic and nonhemolytic. Acid produced from glycerol, glucose, fructose, and mannose but not from erythritol, arabinose, xylose, adonitol, p-methyl-xyloside, galactose, L-sorbose, rhamnose, dulcitol, inositol, mannitol, sorbitol, α-methyl-D-mannoside, α-methyl-D-glucoside, N-acetylglucosamine, amygdalin, arbutin, salicin, maltose, lactose, melibiose, trehalose, melezitose, D-raffinose, glycogen, p-gentiobiose, D-turanose, D-lyxose, D-tagatose, fucose, arabitol, gluconate, and 2-ketogluconate. Acid production from ribose and sucrose variable. Nitrate is not reduced. Urea and esculin are not hydrolyzed. Not lipophilic. CAMP reaction negative. Pyrazinamidase, alkaline phosphatase, esterase, esterase-lipase, cystine arylamidase, and acid phosphatase are positive, but pyrrolidonyl arylamidase and phosphoamidase are variable. Lipase, valine arylamidase, trypsin, chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase activities are not detected.
Cell wall contains meso-diaminopimelic acid. Mycolic acids are present. Propionic acid is not produced as a metabolic product (Bernard et al., 2002).
Source: human disease from various clinical materials.
DNA G+C content (mol%): 63–65 (HPLC).
Type strain: ATCC 700355, CCUG 36878, CIP 105129, DMMZ 2278, DSM 44265, JCM 10384.
Sequence accession no. (16S rRNA gene): Y11200.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492261. By 16S rRNA gene sequencing, Corynebacterium mucifaciens is closely related to (98% identity with) Corynebacterium coyleae and Corynebacterium afermentans, but differs from them (∼94% identity with) when rpoB gene sequences are compared (Khamis et al., 2004).
Corynebacterium mustelae
Funke, Frodl and Bernard 2010b, 873VP
mus.te'la.e. L. n. mustela a weasel, also a scientific zoological name; L. gen. mustelae of a weasel, of Mustela, indicating that the type strain was isolated from a ferret (Mustela putorius furo).
Gram-stain-positive club shaped rods. Unusual greenish-beige adherent colonies with irregular edges, ∼1.0– 2 mm in diameter observed after 48 h. Has odor, described as “humid-cellar like”. Nonlipophilic. Facultatively anaerobic. Acid is produced from ribose, glucose, fructose, mannose, N-acetylglucosamine, arbutin, maltose, sucrose, trehalose, gentiobiose, tagatose, L-fucose, and 5-ketogluconate, but not from glycerol, erythritol, L-arabinose, xylose, adonitol, methyl-β-xyloside, galactose, sorbose, rhamnose, dulcitol, inositol, mannitol, sorbitol, methyl-α-D-mannoside, methyl-α-D-glucoside, amygdalin, salicin, cellobiose, lactose, melibiose, inulin, melezitose, raffinose, starch, glycogen, xylitol, turanose, lyxose, D-fucose, arabitol, gluconate, or 2-ketogluconate. The following enzymes are detected: pyrazinamidase, α-glucosidase, esculinase, esterase, esterase lipase, leucine and cystine arylamidases, and phosphoamidase but nitrate reductase, pyrrolidonyl arylamidase, urease, alkaline and acid phosphatases, lipase, valine arylamidase, trypsin, chymotrypsin, α- and β-galactosidases, β-glucuronidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase are not. CAMP reaction negative.
The cell wall contains meso-DAP and mycolic acids. Cellular fatty acids are consistent with those of the genus, but tuberculostearic acid was not detected. The type strain was susceptible to all antibiotic classes tested.
Source: lung tissue at necropsy of a ferret with lethal sepsis.
DNA G+C content (mol%): not available.
Type strain: 3105, CCUG 57279, DSM 45274.
Sequence accession no. (16S rRNA gene): FJ374773.
Further comments: additional GenBank accession numbers include (partial rpoB gene) FJ467330.
Corynebacterium mycetoides
(ex Castellani 1942) Collins 1983, 438VP (Effective publication: Collins 1982b, 399.) (Corynebacterium mycetoides (Castellani) Ortali and Capocaccia 1956, 490)
my.ce.to'i.des. Gr. n. mukês -êtos fungus; L. suff. -oides (from Gr. suff. -eides from Gr. n. eidos that which is seen, form, shape, figure) ressembling, similar; N.L. adj. mycetoides similar to fungi, referring to ulcers caused by fungi.
Surface colonies on blood or nutrient agar are ∼1 mm in diameter after 2–3 d, circular, convex, entire margin, shiny, and yellow pigmented. Straight to slightly curved Gram-stain-positive rods 1–3 × 0.3–0.5 µm; V-forms and coccoidal forms may be observed. May stain unevenly, and metachromatic granules may be present.
Optimal growth temperature 37°C. Facultatively anaerobic. Glucose is fermented; fructose and trehalose are variable, but maltose, mannose, sucrose, arabinose, lactose, dextrin, cellobiose, galactose, raffinose, ribose, sorbose, xylose, melibiose, glycogen, adonitol, arbutin, inositol, and erythritol are not fermented. Gelatin, cellulose, and esculin not hydrolyzed. Indole, methyl red, and Voges–Proskauer negative. Nitrates not reduced, and urease not produced. Phosphatase positive.
Cell wall contains meso-DAP, with alanine and glutamic acid also being present. Arabinose and galactose are the cell-wall sugars. Corynemycolates (30–36 carbon atoms) present. Significant quantities of unusual, α-alkylbranches with odd numbers of carbon atoms such as C15H31 are detected. Metabolic products are not extant. Cellular fatty acids typical of the genus are detected including the presence of tuberculostearic acid (Bernard et al., 1991). Menaquinones MK-8(H2) with small amounts of MK-9(H2) found. Diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol, and phosphatidylinositol mannoside, as well as unknown glycolipids and some unknown glycolipids and phospholipids are found.
Source: a type of ulcer (tropicaloid ulcer) on the legs of soldiers in the desert regions of North Africa in 1942; not reported in recent literature.
DNA G+C content (mol%): 59 (Tm).
Type strain: ATCC 43995, CCUG 27538, CIP 55.51, DSM 20632, JCM 9388, NBRC 15289, NCTC 9864.
Sequence accession no. (16S rRNA gene): X82066, X84241.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492262.
Corynebacterium phocae
Pascual, Foster, Alvarez and Collins 1998, 603VP
pho'ca'e. L. n. phoca a seal and also a scientific genus name (Phoca); L. gen. n. phocae of a seal, of Phoca, named because the organism was isolated from the common seal, Phoca vitulina.
Cells are Gram-stain-positive short diphtheroids. Colonies are circular, convex, and slightly glistening. The colony diameter is ∼1 mm after 24 h incubation on sheep blood agar at 37°C. Facultatively anaerobic. Acid is produced from D-glucose, D-mannose, and maltose, but not from adonitol, D-arabitol, L-arabitol, D-arabinose, L-arabinose, amygdalin, arbutin, cellobiose, dulcitol, erythritol, D-fucose, L-fucose, D-fructose, galactose, β-gentiobiose, gluconate, glycerol, glycogen, inositol, inulin, 2-ketogluconate, D-lyxose, mannitol, melibiose, methyl α-D-glucoside, methyl α-D-mannoside, β-methylxyloside, D-raffinose, rhamnose, salicin, sorbitol, L-sorbose, starch, D-tagatose, trehalose, D-turanose, xylitol, and xylose. Acid production from galactose, N-acetylglucosamine, lactose, sucrose, and D-raffinose is variable. Nitrate not reduced to nitrite, and urea hydrolysis variable. Esculin and gelatin are not hydrolyzed. Pyrazinamidase, alkaline phosphatase, esterase-lipase, acid phosphatase, and α-glucosidase are detected, but valine arylamidase, chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, esterase, lipase, cystine arylamidase, trypsin, and α-fucosidase activities are not detected. Leucine arylamidase activity variable.
The cell wall contains meso-DAP, and mycolic acids (C30–C34) are present. Cellular fatty acids are consistent with those found for the genus, but tuberculostearic acid is not produced.
Source: the common seal (Phoca vitulina).
DNA G+C content (mol%): 58 (method unknown).
Type strain: M408/89/1, CCUG 38205, CIP 105741, DSM 44612, JCM 12105.
Sequence accession no. (16S rRNA gene): Y10076.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492277. Strain M408/89/l has the characteristics described for the species except that it produces urease and has a negative leucine arylamidase reaction. M408/89/1 produces acid from galactose but not from D-fructose, N-acetylglucosamine, lactose, sucrose, and D-raffinose.
Corynebacterium pilbarense
Aravena-Roman, Spröer, Sträubler, Inglis and Yassin 2010, 1486VP
pil.ba.ren'se. N.L. neut. adj. pilbarense pertaining to Pilbara, Western Australia, the region from which the strain was isolated.
Gram-stain-positive pleomorphic to short rods. Colonies are creamy, circular, and approximately 0.5–2.0 mm in diameter on Columbia blood agar after 24 h incubation at 37°C. Nonhemolytic. Facultatively anaerobic. Nonlipophilic. Esculin, gelatin, and hippurate are not hydrolyzed. Urease and nitrate reduction are negative. Acid is produced from D-glucose, D-ribose, and sucrose. Acid is not produced from L-arabinose, glycogen, inulin, lactose, maltose, mannitol, D-raffinose, sorbitol, trehalose, or D-xylose. Activity for alkaline and acid phosphatases, leucine arylamidase, pyrazinamidase, pyrrolidonyl arylamidase, and naphthol-AS-BI-phosphohydrolase are detected. No activity is detected for arginine dihydrolase, esterase, ester lipase, α-glucosidase, β-glucosidase, α-galactosidase, β-galactosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, α-mannosidase, α-fucosidase, chymotrypsin, trypsin, valine arylamidase, and cystine arylamidase. Acetoin production was negative.
Mycolic acids are present. Long-chain fatty acids are typical for those of the genus, but tuberculostearic acid is absent.
Source: an anaerobic Bactec vial inoculated with ankle aspirate from a man who was thought to be suffering from gout.
DNA G+C content (mol%): not available.
Type strain: IMMIB WACC 658, CCUG 57942, DSM 45350.
Sequence accession no. (16S rRNA gene): FN295567.
Corynebacterium pilosum
Yanagawa and Honda 1978, 209AL
pi.lo'sum. L. neut. adj. pilosum having much hair; intended to mean having many fimbriae (pili).
(This description also contains information from Collins and Cummins, 1986.)
Gram-stain-positive rods, 0.5 × 1.3 µm, arranged as singly, pairs often at angles, or in irregular masses. Metachromatic granules present. Numerous fimbriae or pili observed using electron microscopy. Colonies on nutrient agar and serum agar are cream to pale yellow, entire, circular, opaque, and about 1 mm in diameter after 24 h incubation at 37°C. No hemolysis on sheep, guinea pig, or rabbit blood agar. Facultatively anaerobic. Pellicle and granular sediment formed in broth. No growth at 5°C or 41.5°C, but cells remain viable for 30 min at 56°C. Complex amino acid requirements. Positive for glucose, fructose, mannose, maltose, trehalose, dextrin, and starch; negative for arabinose, xylose, rhamnose, galactose, lactose, sucrose, raffinose, and salicin. Nitrate is reduced, urease is produced, hippurate is hydrolyzed, and pyrazinamidase is detected. Esculin, tyrosine, and gelatin are not hydrolyzed; casein is not digested; methyl red and phosphatase are negative.
meso-DAP and the sugars arabinose and galactose are present. Short-chain mycolates are present (De Briel et al., 1992). Menaquinone MK-8(H2) detected. Cellular fatty acids are consistent with those found for the genus, and tuberculostearic acid is detected (Bernard et al., 1991).
Source: the urine and vagina of healthy cows, but occasionally causes cystitis and pyelonephritis.
DNA G+C content (mol%): 57.9–60.9 (Tm).
Type strain: 46 Hara, ATCC 29592, CCUG 27193, CIP 103422, DSM 20521, JCM 3714, NBRC 15285, NCTC 11862.
Sequence accession no. (16S rRNA gene): X81908, X84246.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492258.
Corynebacterium propinquum
Riegel, de Briel, Prévost, Jehl and Monteil 1994a, 370VP (Effective publication: Riegel, de Briel, Prévost, Jehl and Monteil 1993a, 232.)
pro.pin'qu.um. L. neut. adj. propinquum near, close.
Gram-stain-positive with occasional metachromatic granules and arranged in palisades or V-shaped forms. Colonies (1–2 mm in diameter) on 5% sheep blood agar, nonhemolytic, gray-white with a matted surface. Facultatively anaerobic. Characteristically “ANF”, i.e. an absolute nonfermenter, but reduces nitrate to nitrite. Acid is not produced from D-glucose, glycogen, lactose, sucrose, ribose, D-xylose, D-mannose, D-galactose, trehalose, and D-mannitol or on triple-sugar iron agar. Tyrosine degraded but gelatin, starch, and urea are not. Esculin not hydrolyzed. The methyl red test is negative; acetoin not produced. Growth is visible in 6.5% NaCl. Hydrolysis of hippurate is variable. Production of pyrazinamidase, pyrrolidonyl arylamidase, alkaline phosphatase is variable, but β-glucuronidase, β-galactosidase, α-glucosidase, N-acetyl-glucosaminidase are not produced. Cells utilize acetate, lactate, glyconate, and malate. Utilization of adipate and phenyl acetate is variable. Propionate, glucose, arabinose, mannose, mannitol, N-acetylglucosamine, maltose, caprate, and citrate are not utilized.
Cell wall contains meso-DAP and the sugars arabinose and galactose. The predominant types of mycolic acids are isomers with 30–36 carbons. Cellular fatty acids are consistent with those of the genus.
Source: human sources, mainly from the respiratory tract, but also from other sites.
DNA G+C content (mol%): 57–59 (HPLC).
Type strain: B 77159, ATCC 51488, CCUG 33048, CIP 103792, DSM 44285, JCM 12106.
Sequence accession no. (16S rRNA gene): X81917, X84438.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492279.
Corynebacterium propinquum was formerly called Corynebacterium group ANF-3. Species has <2% variance by 16S rRNA gene sequencing analysis from Corynebacterium pseudodiphtheriticum, but may be distinguished by rpoB analysis (Khamis et al., 2004).
Corynebacterium pseudodiphtheriticum
Lehmann and Neumann 1896, 361AL
Historically, this species has been described by a number of synonyms, including “Bacillus pseudodiphtheriticus” (Lehmann and Neumann 1896) Kruse 1886, “Bacterium pseudodiphtheriticum” (Lehmann and Neumann 1896) Migula 1900, “Mycobacterium pseudodiphthericum” (sic) (Lehmann and Neumann 1896) Chester 1901, “Corynebacterium pseudodiphthericum” (sic) (Lehmann and Neumann 1896) Bergey et al. 1925 and had also been referred to as “Corynebacterium hofmannii” Holland 1920, a name which was never validly published.
pseu.do.diph.the.ri'ti.cum. Gr. adj. pseudes false; N.L. fem. n. diphtheria diphtheria; N.L. adj. diphtheriticus -a -um diphtheric; N.L. neut. adj. pseudodiphtheriticum relating to false diphtheria.
(This description is largely based on that of Collins and Cummins, 1986.)
Short (0.5–2.0 × 0.3–0.5 µm), regular Gram-stain-positive rods which stain evenly except for a transverse medial unstained septum; club forms seen. Metachromatic granules absent or minimally observed. Organisms said to lie in rows with long axes parallel. Grows well on all media. When grown on blood agar, colonies are white to cream colored, regular and smooth, butyrous consistency, and are nonhemolytic. Facultatively anaerobic. Characteristically does not attack commonly used carbohydrates, although it can utilize a wide variety of amides, esters, amino acids, and other organic compounds. Organisms reduce nitrate and hydrolyze urea. Hippurate and pyrazinamidase positive but negative for phosphatase. Optimal temperature 37°C.
Cell-wall sugars are arabinose, galactose, and glucose, and contains meso-DAP. Corynemycolates are present. Cellular fatty acids are consistent with those found for the genus (Bernard et al., 1991). Propionic acid is not produced as metabolic product (Bernard et al., 2002).
Source: nasal mucosa and various human clinical materials.
DNA G+C content (mol%): 54.9–56.8 (Tm).
Type strain: ATCC 10700, CCUG 27539, CIP 103420, DSM 44287, JCM 11665, NBRC 15362, NCTC 11136.
Sequence accession no. (16S rRNA gene): AJ439343, X81918, X84258.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492232. Species has <2% variance by 16S rRNA gene sequencing analysis from Corynebacterium propinquum, but may be distinguished by rpoB analysis (Khamis et al., 2004). Although occasionally present in flora of the upper respiratory tract in healthy individuals, this agent has been associated with exudative pharyngitis mimicking diphtheria, occasionally provoking public health response (Izurieta et al., 1997); recognized as cause of pneumonia, endocarditis, lymphadenitis, keratitis, conjunctivitis, and septic arthritis. Diseases caused by this agent and clinical sites of recovery have been reviewed (Camello et al., 2009). Corynebacterium pseudodiphtheriticum strains can be multidrug resistant, which has been linked to the presence of the ermX gene (Olender and Niemcewicz, 2010).
Corynebacterium pseudotuberculosis
(Buchanan 1911) Eberson 1918, 294AL
Synonyms: “Bacillus pseudotuberculosis-ovis” Lehmann and Neumann 1896, “Bacillus pseudotuberculosis” Buchanan 1911, “Corynebacterium ovis” Bergey et al. 1923, “Corynebacterium pseudotuberculosis-ovis” (Lehmann and Neumann 1896) Hauduroy et al. 1937, “Corynebacterium preisz-nocardi” Hauduroy et al. 1937, “Mycobacterium tuberculosis-ovis” Krasil'nikov 1941.
pseu.do.tu.ber.cu.lo'sis. Gr. adj. pseudes false; N.L. fem. n. tuberculosis tuberculosis; N.L. gen. n. pseudotuberculosis of false tuberculosis.
(This description is largely based on that of Collins and Cummins, 1986.)
In stained smears, described as being similar to Corynebacterium diphtheriae, especially the gravis type, with small irregular rods, 0.5–0.6 × 1.0–3.0 µm, with club forms and metachromatic granules. Fimbriae (pili) may be observed. Colonies on blood agar are yellowish-white, opaque, convex with a matt surface, about 1 mm after 24 h incubation at 37°C. Hemolysis may be observed on horse blood agar (Peel et al., 1997). Like Corynebacterium diphtheriae and Corynebacterium ulcerans, Corynebacterium pseudotuberculosis produces cystinase, as demonstrated by production of brown colonies and haloes when grown on modified Tinsdale medium. Facultatively anaerobic. Positive for glucose, fructose, galactose, mannose, and maltose. Urease production and methyl red are positive. Variable for arabinose, sucrose, dextrin, and trehalose; variable for gelatin, nitrate reduction, and starch hydrolysis. Negative for xylose, rhamnose, lactose, raffinose, and salicin. Negative for esculin and hippurate hydrolysis. Neither pyrazinamidase nor phosphatase is detected. Tyrosine is not hydrolyzed, and casein is not digested. Biochemically, this species is most similar to urease-producing Corynebacterium ulcerans but differs by occasionally reducing nitrate and being negative or rarely reactive with starch, glycogen, or trehalose (Dorella et al., 2006; Hollis and Weaver, 1981; Peel et al., 1997).
Cell walls contain arabinose, galactose, glucose, and mannose and meso-DAP. Menaquinone MK-8(H2) is present. Polar lipids detected include diphosphatidylglycerol, phosphatidylinositol, and monoacylated phosphatidylinositol dimannoside. Corynemycolates are present. Cellular fatty acids are those described for the genus except, as for cysteinase-producing species Corynebacterium diphtheriae, Corynebacterium pseudotuberculosis, and Corynebacterium ulcerans, has significant quantities of a C16:1 isomer. Tuberculostearic acid is not detected (Bernard et al., 1991). Propionic acid is detected as a fermentation product (Bernard et al., 2002).
Source: infections in sheep, goats, horses, and other warm-blooded animals; occasionally linked to disease in humans.
DNA G+C content (mol%): 51.8–52.5 (Tm).
Type strain: ATCC 19410, CCUG 2806, CIP 102968, DSM 20689, JCM 9389, NBRC 15363, NCTC 3450.
Sequence accession no. (16S rRNA gene): X81916, X84255.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492239. Corynebacterium pseudotuberculosis and Corynebacterium ulcerans are the only Corynebacterium species to produce phospholipase D (PLD), with expression of the pld gene product being detected by inhibition of the CAMP reaction (Barksdale et al., 1981). PLD is believed to be a major virulence determinant for spread of caseous lymphadenitis, a disease primarily of sheep and goats. PLD spreads the bacterium from the site of infection to the lymph nodes followed by caseation by means of a complex regulatory process (McKean et al., 2007). Disease in humans is usually occupationally linked to those who handle infected animals (McKean et al., 2007; Peel et al., 1997). Detection of the pld gene has been used clinically to diagnose diseased animals (Pacheco et al., 2007). Corynebacterium pseudotuberculosis isolates may produce diphtheria toxin with the potential to cause diphtherial-like disease, which clinically should be tested for (as described elsewhere in this chapter). Nontoxigenic strains otherwise have typical features. By 16S rRNA gene sequencing, Corynebacterium pseudotuberculosis is most closely related to (>99% identity with) Corynebacterium ulcerans and to (>97% identity with) Corynebacterium diphtheriae, but Corynebacterium ulcerans and Corynebacterium pseudotuberculosis are only distantly related (93% level) when rpoB gene sequences are compared (Khamis et al., 2004).
Corynebacterium pyruviciproducens
Tong, Liu, Summanen, Xu and Finegold 2010, 1139VP
py.ru.vi.ci.pro.du'cens. N.L. n. acidum pyruvicum pyruvic acid; L. part. adj. producens producing; N.L. part. adj. pyruviciproducens pyruvic acid-producing.
Gram-stain-positive coryneform rods. Lipophilic. Can grow aerobically or facultatively anaerobically at 37°C and 42°C. Optimum growth at 37°C under aerobic conditions. Colonies are small, i.e., 0.3–0.5 mm in diameter, circular, entire, convex, and translucent after 2 d of incubation on blood agar. Urease negative. Cannot reduce nitrates. CAMP negative. Acid is produced from D-ribose, D-xylose, D-glucose, maltose, and sucrose, but not from fructose, D-mannitol, lactose, or glycogen. Cells produce esterase, esterase lipase, leucine arylamidase, pyrazinamidase, a small amount of naphthol-AS-BI-phosphohydrolase, and pyrrolidonyl arylamidase, but not acid phosphatase, β-galactosidase, α-glucosidase, N-acetyl-β-glucosaminidase, lipase, valine arylamidase, cystine arylamidase, α-chymotrypsin, α-galactosidase, α-fucosidase, alkaline phosphatase, α-mannosidase, or β-glucosidase. Trypsin and gelatin are not hydrolyzed. β-Lactamase negative. Susceptible to the vibriostatic agent O/129 and highly sensitive to ampicillin, ceftriaxone, clindamycin, and erythromycin.
Cellular fatty acids are consistent for those of the genus, but no tuberculostearic acid is detected. Short-chain mycolic acids (C22–C36) are present. Metabolic products include acetic and pyruvic acids but not propionic.
Source: a human groin abscess.
DNA G+C content (mol%): 62 (HPLC).
Type strain: 06–17730, ATCC BAA-1742, CCUG 57046, WAL 19168.
Sequence accession no. (16S rRNA gene): FJ185225.
Further comments: additional GenBank accession numbers include (partial rpoB gene) FJ899747.
Corynebacterium renale
(Migula 1900) Ernst 1906, 89AL (Bacterium renale Migula 1900, 504)
re.na'le. L. neut. adj. renale pertaining to the kidneys.
(This description is largely based on that of Collins and Cummins, 1986.)
Large, irregularly staining bacillus, 0.7 × 3.0 µm, often with pointed ends; fimbriae (pili) may be present after examination by electron microscopy. Colonies on blood agar are yellow pigmented after 24 h incubation at 37°C. Grows in broth at pH 5.4. Facultatively anaerobic. Positive for glucose, fructose, mannose, and dextrin. Variable for maltose and trehalose. Negative for arabinose, xylose, rhamnose, galactose, lactose, sucrose, raffinose, starch, and salicin. Urease production and methyl red are positive. Hippurate hydrolysis positive. Pyrazinamidase positive, but phosphatase negative. Esculin, tyrosine, and gelatin are not hydrolyzed. Casein is digested. Nitrate is not reduced.
Corynemycolates (30–36 carbons) and menaquinone MK-8(H2) are present. Cellular fatty acids consistent with those for the genus are found, but tuberculostearic acid is not detected. Propionic acid is not detected as fermentation product (Bernard et al., 2002).
Source: cystitis and pyelitis in cattle and occasionally in other animals. Disease in humans described historically, but no genetic confirmation.
DNA G+C content (mol%): 53–58 (Tm).
Type strain: ATCC 19412, CCUG 27542, CIP 103421, DSM 20688, HAMBI 2321, JCM 9391, NBRC 15290, NCTC 7448.
Sequence accession no. (16S rRNA gene): X81909, X84249.
Further comments: additional GenBank accession numbers include (complete rpoB gene) Y492240.
Corynebacterium resistens
Otsuka, Kawamura, Koyama, Iihara, Ohkusu and Ezaki 2005b, 2235VP (Effective publication: Otsuka, Kawamura, Koyama, Iihara, Ohkusu and Ezaki 2005a, 3716.)
re.sis.tens. L. part. adj. resistens enduring, resistant (multidrug-resistant).
Cells are Gram-stain-positive, typically club-shaped rods, 1–3 µm in length, and arranged as single cells, in pairs, or in small clusters. Growth on TSA with 5% sheep blood produces nonpigmented, grayish-white, glistening, pearly colonies up to 1.0 mm in diameter. Colonies are nonhemolytic and very slow-growing under anaerobic conditions. Tween 80 enhances growth, resulting in colonies 2–4 mm in diameter, i.e. strains are lipophilic. CAMP negative. Nitrate not reduced. Ferments D-tagatose, 5-ketogluconate, ribose, and D-glucose with trehalose and L-sorbose being variable. Negative for glycerol, erythritol, D-arabinose, L-arabinose, D-xylose, L-xylose, adonitol, β-methylxyloside, galactose, D-fructose, D-mannose, rhamnose, dulcitol, inositol, mannitol, sorbitol, α-methyl-D-mannoside, α-methyl-D-glucoside, N-acetylglucosamine, amygdalin, arbutin, esculin, salicin, cellobiose, maltose, lactose, melibiose, sucrose, inulin, melezitose, D-raffinose, starch, glycogen, xylitol, β-gentiobiose, D-turanose, D-lyxose, D-fucose, L-fucose, D-arabitol, L-arabitol, gluconate, and 2-ketogluconate. Alkaline phosphatase, esterase, esterase lipase, leucine arylamidase, acid phosphatase, and naphthol-AS-BI-phosphohydrolase strongly positive, but lipase, cysteine, and arylamidase are weakly positive. Reactions for pyrazinamidase, valine arylamidase, trypsin, chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase are negative.
Cellular fatty acids are consistent for those described for the genus; other chemotaxonomic information is not extant.
Source: human disease from bacteremias of leukemic patients, bronchial aspirates of patients with lymphoma or subarachnoid hemorrhaging, and a patient with cellulitis.
DNA G+C content (mol%): 54.643 (standard deviation, 0.03%) (HPLC).
Type strain: CCUG 50093, GTC 2026, JCM 12819, SICGH 158.
Sequence accession no. (16S rRNA gene): AB128981.
Further comments: Corynebacterium resistens is resistant to multiple drug classes, but all strains are susceptible to vancomycin.
Corynebacterium riegelii
Funke, Lawson and Collins 1998b, 627VP (Effective publication: Funke, Lawson and Collins 1998a, 626.)
ri.e.gel'i.i. N.L. gen. masc. n. riegelii of Riegel, to honor contemporary French microbiologist Philippe Riegel for his contributions to the taxonomy of the genus Corynebacterium as well as to the clinical microbiology of coryneform bacteria.
Gram-stain-positive typically club-shaped rods which appear as single cells, in pairs, or in small clusters. Colonies are whitish with a creamy or a slightly sticky consistency, circular with entire edges, convex, glistening, and up to 1.5 mm in diameter after 48 h of incubation. Weak growth anaerobically. Biochemically unusual, as acid is produced from maltose, ribose, trehalose, D-tagatose, and 5-ketogluconate, but not produced from glucose, sucrose, mannitol, xylose, glycerol, erythritol, arabinose, adonitol, β-methylxyloside, galactose, D-fructose, D-mannose, L-sorbose, rhamnose, dulcitol, inositol, sorbitol, α-methyl-D-mannoside, α-methyl-D-glucoside, N-acetylglucosamine, amygdalin, arbutin, salicin, cellobiose, lactose, melibiose, inulin, melezitose, D-raffinose, starch, glycogen, xylitol, β-gentiobiose, D-turanose, D-lyxose, fucose, arabitol, gluconate, or 2-ketogluconate. Nitrate is not reduced. Urea hydrolysis strongly positive, but esculin not hydrolyzed. CAMP reaction negative. Esterase, esterase lipase, leucine arylamidase, and cystine arylamidase detected, but pyrazinamidase and alkaline and acid phosphatases are variable. Pyrrolidonylarylamidase, lipase, valine arylamidase, trypsin, chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase are not detected. The type strain has features as described above except that the activity of alkaline phosphatase but not that of pyrazinamidase or acid phosphatase is detected.
The cell wall contains meso-DAP. Mycolic acids are present. Cellular fatty acids are consistent with those described for the genus, but tuberculostearic acid is not detected. Propionic acid is not produced as a metabolic product (Bernard et al., 2002).
Source: human clinical specimens.
DNA G+C content (mol%): not available.
Type strain: ATCC 700782, CCUG 38180, CIP 105310, DMMZ 2415, DSM 44326, JCM 10389.
Sequence accession no. (16S rRNA gene): Y14651.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492278.
Corynebacterium simulans
Wattiau, Janssens and Wauters 2000, 351VP
si'mu.lans. L. part. adj. simulans simulating, because it resembles Corynebacterium striatum.
Cells are Gram-stain-positive showing a diphtheroid arrangement. Colonies are grayish-white, glistening, and 1–2 mm in diameter on blood agar after 48 h incubation at 37°C. Facultatively anaerobic. Fermentative. Growth does not occur or is very weak at 20°C within 3 d. Acid is produced from glucose, sucrose, fructose, and mannose. Acid production from N-acetylglucosamine, galactose, and ribose is variable. Acid is not produced from mannitol, maltose, D- and L-xylose, glycerol, erythritol, D- and L-arabinose, adonitol, methyl-β-D-xyloside, sorbose, rhamnose, dulcitol, inositol, sorbitol, methyl-α,D-mannoside, methyl-α,D-glucoside, amygdalin, arbutin, esculin, salicin, cellobiose, lactose, melibiose, trehalose, inulin, melezitose, raffinose, starch, glycogen, xylitol, gentiobiose, D-turanose, D-lyxose, D-tagatose, D- and L-fucose, D- and L-arabitol, gluconate, 2-ketogluconate, and 5-keto-gluconate. Urea, esculin, and gelatin not hydrolyzed. Nitrate and nitrite reduction positive, a feature unique among all Corynebacterium species with nitrite reduction occurring at concentrations of 0.001–0.01%.
Alkalinization of buffered formate is positive. No acid produced from ethylene glycol. Tween esterase and tyrosine clearing positive. Pyrazinamidase is variable. CAMP reaction negative. Alkaline phosphatase, esterase, esterase-lipase, leucine arylamidase, trypsin, and naphthol-AS-BI-phosphohydrolase positive but lipase, valine arylamidase, cystine arylamidase, chymotrypsin, acid phosphatase, α- and β-galactosidase, β-glucuronidase, α- and β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase negative. The type strain of Corynebacterium simulans is as described here with ribose, N-acetylglucosamine, galactose, and pyrazinamidase being positive. Cell wall contains the sugars arabinose and galactose as well as meso-DAP. Cellular fatty acids are consistent with those described for the genus. Short-chain mycolic acids (C22–C36) are present. Propionic acid is not detected as a metabolic product (Bernard et al., 2002).
Source: human clinical materials including bile (Bernard et al., 2002; Wattiau et al., 2000).
DNA G+C content (mol%): not available.
Type strain: Co 553, ATCC BAA-15, CCUG 43305, CIP 106488, DSM 44415, JCM 12107, UCL 553.
Sequence accession no. (16S rRNA gene): AJ012837.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492264.
Corynebacterium singulare
Riegel, Ruimy, Renaud, Freney, Prévost, Jehl, Christen and Monteil 1997b, 1095VP
sin.gu.la're. L. neut. adj. singulare single, unique.
Gram-stain-positive irregular rods arranged in typical V-shaped forms or palisades. Facultatively anaerobic. Non-hemolytic colonies (1–2 mm diameter) grayish and smooth after 24 h at 37°C on sheep blood agar. Colonies circular and slightly convex with entire margins. Nitrate is not reduced to nitrite. Urea and tyrosine degraded, but gelatin and esculin are not hydrolyzed. Acid is produced from glucose, maltose, and sucrose but not from lactose, glycogen, ribose, trehalose, mannitol, and D-xylose. Alkaline phosphatase, pyrrolidonylarylamidase, and pyrazinamidase are produced but α-glucosidase, β-glucuronidase, β-galactosidase, and N-acetyl-β-glucosaminidase are not. Cells utilize D-glucose, D-fructose, D-trehalose, D-mannose, sucrose, maltose, glycerol, D-turanose, L-malate, phenylacetate, putrescine, DL-lactate, caprylate, L-histidine, succinate, fumarate, 3-hydroxybutyrate, L-aspartate, L-glutamate, D-alanine, L-serine, propionate, and L-tyrosine as sole carbon sources. D-Ribose, N-acetyl-D-glucosamine, and D-gluconate are not utilized.
Cell wall contains meso-DAP, the sugars arabinose and galactose, and short-chain mycolic acids (C26–C36). Cellular fatty acids are consistent with those described for the genus. Propionic acid is not produced by anaerobic metabolism of glucose.
Source: human semen.
DNA G+C content (mol%): 62 (capillary electrophoresis).
Type strain: CCUG 37330, CIP 105491, DSM 44357, IBS B52218, JCM 10385, NBRC 16162.
Sequence accession no. (16S rRNA gene): Y10999.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492280.
By 16S rRNA gene sequencing alone, this species can not be readily discerned from Corynebacterium aurimucosum and Corynebacterium minutissimum, but these can be resolved using rpoB gene sequencing (Khamis et al., 2004).
Corynebacterium sphenisci
Goyache, Ballesteros, Vela, Collins, Briones, Hutson, Potti, García-Borboroglu, Domínguez and Fernández-Garayzábal 2003a, 1011VP
sphe.nis'ci. N.L. masc. gen. n. sphenisci of Spheniscus, a systematic genus of penguins, source of the type strain.
Gram-stain-positive rods. Colonies are whitish, low-convex, dry and rough, ∼1–2 mm in diameter after 48 h incubation at 37°C on sheep blood agar. Facultatively anaerobic. Nonhemolytic, nonlipophilic, and CAMP negative. Nitrates are reduced. Acid is produced from glucose, maltose, galactose, fructose, mannose, and trehalose, but not from ribose, sucrose, glycogen, xylose, mannitol, lactose, erythritol, D-arabinose, L-arabinose, adonitol, galactose, L-sorbose, rhamnose, inositol, sorbitol, methyl-α-D-mannoside, methyl-α-D-glucoside, amygdalin, dulcitol, arbutin, salicin, cellobiose, melibiose, inulin, melezitose, D-raffinose, xylitol, β-gentiobiose, D-turanose, D-lyxose, D-tagatose, D-fucose, L-fucose, D-arabitol, L-arabitol, N-acetylglucosamine, 2-ketogluconate, 5-ketogluconate, or glycerol. Gelatin, urea, and esculin are not hydrolyzed. Pyrazinamidase, esterase, ester lipase, lipase, leucine arylamidase, valine arylamidase, cystine arylamidase, chymotrypsin, acid phosphatase, and naphthol-AS-BI-phosphohydrolase detected, but alkaline phosphatase, pyrrolidonyl arylamidase, α-glucosidase, β-glucosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, α-mannosidase, α-galactosidase, β-galactosidase, α-fucosidase, and trypsin are not.
Cell-wall murein is based on meso-DAP. Cellular fatty acids are consistent with those described for the genus, but tuberculostearic acid is not present. Corynomycolates detected in small amounts.
Source: the cloaca of an apparently healthy wild magellanic penguin (Spheniscus magellanicus).
DNA G+C content (mol%): not available.
Type strain: CCUG 46398, CECT 5990, JCM 12270.
Sequence accession no. (16S rRNA gene): AJ440964.
Further comments: additional GenBank accession numbers include (partial rpoB gene) EU004067 for DSM 44792.
Corynebacterium spheniscorum
Goyache, Vela, Collins, Ballesteros, Briones, Moreno, Yorio, Domínguez, Hutson and Fernández-Garayzábal 2003b, 45VP
sphe.nis.co'rum. N.L. masc. n. spheniscus a genus of penguin; N.L. masc. pl. gen. n. spheniscorum of penguins.
Gram-stain-positive rods. Colonies are whitish, circular, smooth, entire, and ∼ 1–2 mm diameter on Columbia blood agar after 24 h incubation at 37°C. Nonhemolytic.
Facultatively anaerobic; grows slightly under anaerobic conditions. Nonlipophilic, and CAMP-positive using Staphylococcus aureus in the test assay. Esculin, gelatin, and urea are not hydrolyzed. Nitrate not reduced. Acid is produced from glucose, ribose, D-fructose, D-mannose, trehalose, N-acetyl-β-glucosamine, and maltose, but not from D-xylose, L-xylose, mannitol, lactose, rhamnose, galactose, adonitol, inositol, D- or L-arabinose, sucrose, melibiose, melezitose, or glycogen. Acidification of dulcitol is variable. Enzyme activity for ester lipase, esterase, pyrazinamidase, and leucine arylamidase (weak) detected but not for alkaline and acid phosphatases, pyrrolidonyl arylamidase, α-glucosidase, β-glucosidase, β-glucuronidase, α-galactosidase, β-galactosidase, α-mannosidase, α-fucosidase, chymotrypsin, trypsin, valine arylamidase, cystine arylamidase, and naphthol-AS-BI-phosphohydrolase.
Cell wall contains meso-DAP, and Cellular fatty acids are consistent with those described for the genus, but tuberculostearic acid is not present. Mycolic acids are present (C32–C36).
Source: the cloacae of apparently healthy penguins (Spheniscus magellanicus).
DNA G+C content (mol%): not available.
Type strain: PG 39, CCUG 45512, CECT 5986, JCM 12271.
Sequence accession no. (16S rRNA gene): AJ429234.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492283.
Corynebacterium sputi
Yassin and Siering 2008, 2878VP
spu'ti. L. gen. n. sputi of sputum.
Gram-stain-positive rods. Colonies are cream-yellow, circular, convex, dry, and ∼ 0.2–2 mm in diameter on Columbia blood agar after 48 h incubation at 37°C. Nonhemolytic. Lipophilic, as Tween 80 encourages growth of the organism. Facultatively anaerobic. Urea hydrolyzed, but esculin, gelatin, and hippurate are not. Nitrate not reduced. Acid produced from D-glucose but not from L-arabinose, glycogen, inulin, D-lactose, maltose, D-mannitol, raffinose, D-ribose, D-sorbitol, starch, sucrose, trehalose, or D-xylose. Activity detected for α-glucosidase, lipase, lipase, leucine arylamidase, pyrazinamidase, and naphthol-AS-BI-phosphohydrolase but not for acid and alkaline phosphatases, arginine dihydrolase, chymotrypsin, cysteine arylamidase, esterase, α-fucosidase, α-galactosidase, β-galactosidase, β-glucuronidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, pyrrolidonyl arylamidase, trypsin, or valine arylamidase. Acetoin production negative.
Corynemycolic acids are present. Cellular fatty acids are consistent with those described for the genus, and tuberculostearic acid is present.
Source: the sputum of a patient with pneumonia.
DNA G+C content (mol%): not available.
Type strain: IMMIB L-999, CCUG 55795, DSM 45148.
Sequence accession no. (16S rRNA gene): AM930556.
Corynebacterium stationis
(ZoBell and Upham 1944) Bernard, Wiebe, Burdz, Reimer, Ng, Singh, Schindle and Pacheco 2010, 877VP (“Achromobacter stationis” Zobell and Upham 1944, 273; Brevibacterium stationis Breed 1953, 14)
sta.ti.o'nis. L. gen. n. stationis of a fixed position.
Gram-stain-positive short rods, 0.6–1.0 µm in diameter, occurring singly, in pairs, in “V” forms, and can be club shaped. Colonies on blood agar after 24 h are gray-white but become yellowish with age, ∼1 mm in diameter, raised, with no hemolysis observed. Facultatively anaerobic, but grows better under aerobic conditions. Poor or no growth under strictly anaerobic conditions. Grows at 25, 35, and 42°C in air. Nonlipophilic. Acid is produced, albeit slowly, from glucose, fructose, and ribose. Mannose and fucose found in the API 50CH gallery also weakly reactive. Xylose, mannitol, lactose, sucrose, maltose, galactose, glycerol, glycogen, raffinose, salicin, and trehalose are not reactive. Triple-sugar iron remains neutral or alkaline/neutral. Grows in the presence of 6–10% NaCl. Urease produced. Nitrate reduced to nitrite, but nitrite is not reduced to nitrogen using either conventional or API Coryne panel methods. Simmons' citrate is alkalinized. Tyrosine is hydrolyzed, but gelatin, esculin, casein, and starch are not. Not reactive in tests for lysine, arginine, or ornithine decarboxylases. CAMP and CAMP-inhibition reactions not observed. DNase and indole are not produced. Pyrazinamidase may be produced, but otherwise most enzymes of the API ZYM panel are not detected. Strains are generally susceptible to a wide variety of antimicrobials.
Cell wall contains meso-DAP and the sugars are arabinose and galactose. Corynemycolates are present. Menaquinones MK-8(H2) and MK-9(H2) detected. Cellular fatty acids are consistent with those described for the genus. Propionic acid is not detected as metabolic product.
Source: sea water. ATCC 6872, previously identified as Corynebacterium ammoniagenes but now assigned to Corynebacterium stationis, was isolated from human infant stools. Additional strains were recovered from human blood cultures (Bernard et al., 2010).
DNA G+C content (mol%): 53.9 (Tm).
Type strain: ATCC 14403, CCUG 43497, CIP 104228, DSM 20302, JCM 11611, NBRC 12144, VKM B-1228.
Sequence accession no. (16S rRNA gene): FJ172667.
Further comments: additional GenBank accession numbers include (partial rpoB gene) FJ172671.
Corynebacterium striatum
(Chester 1901) Eberson 1918, 22AL (Bacterium striatum Chester 1901, 171)
stri.a'tum. L. part. neut. adj. striatum grooved.
Note: None of the reference strains contained in two major culture collections (ATCC, NCTC) fit the descriptions found in the original publications and the wrong strains appear to have been deposited for this species. Therefore, there is a case for declaring Corynebacterium striatum a nomen dubium (Rule 56a) exists. However, if a careful search reveals a strain or strains used in the original description agreeing in its characteristics, then a neotype strain can be proposed (Rule 18c). The description here is based primarily on characteristics (now widely accepted for this species) linked to reactions for ATCC 6940 (Coyle et al., 1993a; Funke and Bernard, 2007; Hollis and Weaver, 1981; Leonard et al., 1994; Martinez-Martinez et al., 1995b). The description shown here outlines several biochemical differences from those outlined in the previous edition of this Manual (Collins and Cummins, 1986).
Pleomorphic Gram-stain-positive rods, often club-shaped, 0.25–0.5 × 2.0–3.0 µm. Coccoidal forms and long filaments can be found among older cultures. Metachromatic granules may be observed. Colonies may be slow-growing, white, smooth, entire, about 1 mm in diameter after 48 h of incubation at 37°C on sheep blood agar (Collins), but some are creamy, pale-yellow colonies. A strain which caused multiple cases of pneumonia in an ICU was brown pigmented (Leonard et al., 1994). Nonhemolytic, but slight hemolysis may be seen around deep cultures. Nonlipophilic. Facultatively anaerobic. Positive for glucose, fructose, mannose, starch, and dextrin. Variable for galactose and trehalose. Nitrate is reduced, maltose and lactose are negative, and reaction for sucrose is variable. Arabinose, xylose, mannitol, rhamnose, raffinose, and salicin are negative. This is in contrast to a negative reaction for nitrate reduction, a positive reaction for maltose fermentation, and variable reactions for lactose and sucrose as described in the previous version of Bergey's Manual (Collins and Cummins, 1986). Urease is not produced, and methyl red is variable (Coyle et al., 1993a). Pyrazinamidase and alkaline phosphatase positive. Esculin not hydrolyzed. Gelatin is described as variable (Collins and Cummins, 1986) or negative (Coyle et al., 1993a). Tyrosine hydrolyzed (Funke and Bernard, 2007; Martinez-Martinez et al., 1995b).
ATCC 6940T cell wall contains arabinose and galactose as well as mycolic acids. Menaquinone MK-8(H2) detected. Cellular fatty acids are like those described for the genus, but tuberculostearic acid is not detected (Bernard et al., 1991). Propionic acid is not detected as a metabolic product of glucose fermentation (Bernard et al., 2002).
Source: human nasopharynx; strains recovered from a variety of human materials, but historically also recovered from the milk of cows with mastitis.
DNA G+C content (mol%): 57.6 (Tm).
Type strain: ATCC 6940, CCUG 27949, CIP 81.15, DSM 20668, JCM 9390, NBRC 15291, NCTC 764.
Sequence accession no. (16S rRNA gene): X81910, X84442.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492267. Multidrug resistant strains have been increasingly documented among patients with Corynebacterium striatum infections (Tarr et al., 2003) and it may cause potentially fatal disease among compromised patients (Lee et al., 2005; Otsuka et al., 2006). Person to person transmissions among ICU patients have been documented (Brandenburg et al., 1996; Leonard et al., 1994). Also associated with bovine mastitis (Coyle et al., 1993a).
Corynebacterium suicordis
Vela, Mateos, Collins, Briones, Hutson, Domínguez and Fernández-Garayzábal 2003, 2029VP
su.i.cor'dis. L. n. sus suis pig; L. n. cor cordis heart; N.L. gen. n. suicordis of/from pig heart.
Gram-stain-positive rods. Colonies are whitish, circular, smooth, entire, and 1–2 mm in diameter after 48 h incubation at 37°C on sheep blood agar. Facultatively anaerobic. Nonhemolytic. CAMP negative and nonlipophilic. Nitrate is not reduced. Acid is not produced from D-glucose, maltose, lactose, ribose, sucrose, glycogen, mannitol, glycerol, erythritol, D-arabinose, L-arabinose, D-xylose, L-xylose, adonitol, inositol, methyl-β-xyloside, galactose, D-fructose, D-mannose, L-sorbose, rhamnose, methyl-α-D-mannoside, methyl α-D-glucoside, sorbitol, N-acetyl-β-glucosamine, amygdalin, arbutin, salicin, cellobiose, melibiose, trehalose, inulin, melezitose, D-raffinose, xylitol, b-gentiobiose, D-turanose, D-lyxose, D-tagatose, D-fucose, L-fucose, D-arabitol, L-arabitol, 5-ketogluconate, or 2-ketogluconate. Urea hydrolyzed, but esculin and gelatin are not. Pyrazinamidase, esterase, ester lipase, alkaline phosphatase, acid phosphatase, and naphthol-AS-BI-phosphohydrolase detected, but pyrrolidonyl arylamidase, β-glucuronidase, β-galactosidase, α-glucosidase, lipase, leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, chymotrypsin, α-galactosidase, β-glucosidase, α-mannosidase, and α-fucosidase, are not.
Cell wall contains meso-DAP, and corynomycolic acids are present (C28–C36). Cellular fatty acids are consistent with those described for the genus.
Source: the heart, pleural cavity, lymph nodes, and lungs of diseased pigs.
DNA G+C content (mol%): not available.
Type strain: P81/02, CCUG 46963, CECT 5724, JCM 12370.
Sequence accession no. (16S rRNA gene): AJ504424.
Further comments: additional GenBank accession numbers include (partial rpoB gene) EU004068 for CIP 108201.
Corynebacterium sundsvallense
Collins, Bernard, Hutson, Sjödén, Nyberg and Falsen 1999a, 364VP
sunds.val.len'se. N.L. neut. adj. sundsvallense of or belonging to Sundsvall, Sweden, named after the city from where the bacterium was first isolated.
Gram-stain-positive rods; some branching and bulges/knobs at the ends of some cells may be observed. Colonies are buff or yellowish, opaque, shiny, heaped, and adherent to medium. Nonhemolytic. Nonlipophilic and CAMP negative. Growth possible in 6% NaCl but not in 10% NaCl. Acid produced from fructose, glucose, maltose, and sucrose but not from amygdalin, N-acetylglucosamine, galactose, glycogen, lactose, mannitol, ribose, raffinose, salicin, trehalose, or D-xylose. Hippurate is hydrolyzed but esculin, gelatin, and starch are not. Nitrate not reduced. Urease positive and phosphoamidase weakly detected. Variable for α-glucosidase, as 2 strains were positive but 2 are negative (Bernard et al., 2002). Some strains are positive for alkaline phosphatase, leucine arylamidase, pyrazinamidase, ester lipase, and esterase. Acid phosphatase, cystine arylamidase, chymotrypsin, α-fucosidase, α-galactosidase, β-galactosidase, β-glucosidase, β-glucuronidase, lipase, α-mannosidase, pyrrolidonyl arylamidase, trypsin, and valine arylamidase are not detected. Type strain has features as described above except weak reactions for pyrazinamidase and alkaline phosphatase are observed and leucine arylamidase, ester lipase, and esterase activities are not detected.
The cell wall contains meso-DAP, and mycolic acids are present. Cellular fatty acids consistent with those described for the genus, but tuberculostearic acid is not present. Lactate and succinate but not propionate are major products of glucose fermentation.
Source: human clinical specimens.
DNA G+C content (mol%): 64 (Tm).
Type strain: CCUG 36622, CIP 105936, DSM 44613, JCM 12401.
Sequence accession no. (16S rRNA gene): Y09655.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492268. By 16S rRNA gene sequencing, closely related to (∼99.0% identity) to Corynebacterium thomssenii, but differs from that species (∼90.0% identity) when complete sequences of the rpoB gene are compared (Khamis et al., 2004).
Corynebacterium terpenotabidum
Takeuchi, Sakane, Nihira, Yamada and Imai 1999, 228VP
ter.pen.o.ta'bi.dum. N.L. n. terpenum terpene; L. neut. adj. tabidum dissolving; N.L. neut. adj. terpenotabidum terpene-dissolving.
Gram-stain-positive irregular rods, 0.5–0.7 × 19–15 µm long in young cultures; some cells are arranged at a V angle. In older cultures, cells are 0.5–0.7 × 0.6–1.0 µm. Colonies on PY-BHI agar are circular, have a rough surface, and are grayish white. Growth occurs under aerobic, but not anaerobic, conditions. Methyl red test negative. Nitrate not reduced to nitrite. Hugh–Leifson's test in glucose oxidation-fermentation (OF) medium is oxidative. Voges–Proskauer test positive. Urease produced, but arginine dihydrolase, lysine decarboxylase, and ornithine decarboxylase are not. Tween 80 is hydrolyzed, but starch, gelatin, casein, cellulose, esculin, and tyrosine are not. Fructose, glucose, mannose, lactate, and ethanol are utilized as carbon sources, but galactose, lactose, maltose, sucrose, glycerol, sorbitol, mannitol, inositol, citrate, succinate, malonate, pimelate, m-hydroxybenzoate, p-hydroxybenzoate, arginine, aspartate, histidine, methylamine, ethylamine, and methanol are not. Thiamin and biotin required for growth. Growth is observed in PY-BHI broth containing 8% but not 10% NaCl.
Cell wall contains meso-DAP with arabinose, galactose, and mannose, as major cell-wall sugars. The muramic acids of peptidoglycan occur in the N-acetyl form. Menaquinone MK-9(H2) predominates. Mycolic acids are present. Cellular fatty acids are consistent with those described for the genus, and high levels of tuberculostearic acid are present.
Source: soil.
DNA G+C content (mol%): 67.5 (HPLC).
Type strain: Y −11, CIP 105927, JCM 10555, NBRC 14764, VKM Ac-2071.
Sequence accession no. (16S rRNA gene): AB004730.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492269.
Corynebacterium testudinoris
Collins, Hoyles, Hutson, Foster and Falsen 2001b, 1351VP
tes.tu.din.o'ris. L. n. testudo -inis tortoise; L. gen. neut. n. oris of the mouth; N. L. gen. neut. n. testudinoris of the mouth of a tortoise.
Gram-stain-positive rods. Yellow pigmented. Facultatively anaerobic. Nonlipophilic. Acid is produced from glucose, maltose, ribose, and sucrose but not from glycogen, lactose, mannitol, or D-xylose. Activity detected for acid phosphatase (weak), esterase (weak), ester lipase (weak), β-glucosidase, and leucine arylamidase. No activity detected for alkaline phosphatase, cystine arylamidase, chymotrypsin, α-fucosidase, α-galactosidase, β-galactosidase, β-glucuronidase, N-acetyl- β-glucosaminidase, lipase, α-mannosidase, valine arylamidase, pyrazinamidase, phosphoamidase, pyrrolidonyl arylamidase, urease, or trypsin. α-Glucosidase variable. Esculin is hydrolyzed, but gelatin is not. Nitrate is reduced.
Cell wall contains meso-DAP. Cellular fatty acids are consistent with those described for the genus, but tuberculostearic acid is not present. Mycolic acids are present (C30–C36).
Source: necrotic lesions in mouth of a tortoise; habitat is unknown.
DNA G+C content (mol%): not available.
Type strain: M935/96/4, CCUG 41823, CIP 106763, DSM 44614, JCM 12108.
Sequence accession no. (16S rRNA gene): AJ295841.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492284.
Corynebacterium thomssenii
Zimmermann, Spröer, Kroppenstedt, Fuchs, Köchel and Funke 1998, 491VP
thoms.se'ni.i. N.L. gen. masc. n. thomssenii of Thomssen, to honor Reiner Thomssen, a prominent German virologist and medical microbiologist.
Gram-stain-positive diphtheroid-like rods. Colonies of this fastidious slow-growing bacterium are whitish, circular, mucoid, and sticky. After 24 h incubation on Columbia agar supplemented with 5% sheep blood, the colony diameter is less than 0 ± 5 mm. Fermentative. Acid is produced from glucose, maltose, sucrose, D-fructose, D-mannose, trehalose, and 5-keto-gluconate, but not from mannitol, xylose, glycerol, erythritol, arabinose, adonitol, β-methyl-xyloside, L-sorbose, rhamnose, dulcitol, inositol, sorbitol, α-methyl-D-mannoside, α-methyl-D-glucoside, N-acetyl-β-glucosamine, amygdalin, arbutin, salicin, cellobiose, melibiose, inulin, melezitose, D-raffinose, glycogen, xylitol, β-gentiobiose, D-lyxose, L-arabitol, gluconate, and 2-keto-gluconate. Pyrazinamidase, alkaline phosphatase, esterase, esterase lipase, leucine arylamidase, cystine arylamidase, acid phosphatase, and N-acetyl-β-glucosaminidase present, but lipase, valine arylamidase, chymotrypsin, α-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, α-mannosidase, and α-fucosidase are not. One isolate was N-acetyl-β-glucosamine negative (Bernard et al., 2002). Urea hydrolyzed, but nitrate not reduced. Esculin hydrolysis negative. Nonlipophilic and CAMP reaction negative. DNase activity observed after 48 h. Tyrosine not hydrolyzed.
Cell wall contains arabinose and galactose as well as meso-DAP. Corynemycolates (C32–C36) are present. Cellular fatty acids are consistent with those described for the genus. Polar lipids are composed of diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol, and phosphatidylinositol mannosides. MK-8(H2) and MK-9(H2) found; small amounts of MK-10(H2) also found. Propionic acid not detected as a fermentation product (Bernard et al., 2002).
Source: pleural fluid from a patient with chronic renal failure, stroke, and pneumonia and from air (Bernard et al., 2002).
DNA G+C content (mol%): not available.
Type strain: CCUG 38516, CIP 105597, DSM 44276, JCM 12109.
Sequence accession no. (16S rRNA gene): AF010474.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492270. By 16S rRNA gene sequencing, closely related to (∼99.0% identity) Corynebacterium sundsvallense, but differs from that species (∼90.0% identity) when sequences of the rpoB gene are compared (Khamis et al., 2004).
Corynebacterium timonense
Merhej, Falsen, Raoult and Roux 2009, 1956VP
ti.mo.nen'se. N.L. neut. adj. timonense of or pertaining to Hôpital de la Timone, the name of a hospital in Marseille France where the type strain was isolated.
Gram-stain-positive typically club-shaped rods that occur as single cells, in pairs, or in small clusters. Rods are 0.6–2.1 × 0.4–0.6 µm after 48 h growth in TSB medium (as determined by electron microscopy). After 24 h growth on sheep blood agar at 37°C, surface colonies are circular, yellow, shiny, and 1–2 mm in diameter. Capable of aerobic and anaerobic growth. Temperature range for growth is 25–50°C (optimum, 37°C). With the API Coryne system, acid produced from D-glucose and D-ribose, but not from D-xylose, D-mannitol, maltose, D-lactose, sucrose, or glycogen. Nitrate not reduced. Esculin is weakly hydrolyzed, but urea and gelatin are not. With API Coryne and API ZYM strips, positive for pyrazinamidase, alkaline phosphatase, esterase, esterase lipase, lipase (weakly), leucine arylamidase, valine arylamidase, and acid phosphatase, but negative for pyrrolidonyl arylamidase, cystine arylamidase, trypsin, achymotrypsin, naphthol-AS-BI-phosphohydrolase, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase.
Cellular fatty acids are found to be consistent with those described for the genus, but tuberculostearic acid is not detected. Other chemotaxonomic properties were not extant.
Source: multiple blood cultures of a septic patient post-surgically after a pacemaker had been implanted.
DNA G+C content (mol%): not available.
Type strain: 5401744, CCUG 53856, CIP 109424, CSUR P20.
Sequence accession no. (16S rRNA gene): EF217055.
Further comments: additional GenBank accession numbers include (partial rpoB gene) EF217058.
Corynebacterium tuberculostearicum
Feurer, Clermont, Bimet, Candréa, Jackson, Glaser, Bizet and Dauga 2004, 1059VP (“Corynebacterium tuberculostearicum” Brown, Lanéelle, Asselineau and Barksdale 1984, 251)
tu.ber.cu.lo.ste.a'ri.cum. N.L. neut. adj. tuberculostearicum pertaining to tuberculostearic acid which is contained in the cells.
Pleomorphic Gram-stain-positive rods which can develop coccoid forms in stationary cultures. Colonies on trypto casein soy agar supplemented with Tween 80 are circular, convex, glistening, and 1 mm in diameter. Aerobic to facultatively anaerobic. Lipophilic. Acid is produced from galactose, glucose, glycerol, fructose, mannose, ribose, and 5-ketogluconate but not from mannitol, glycogen, starch, sorbitol, lactose, inulin, or xylose. Production of acid from trehalose, maltose, gluconate, sucrose, and N-acetylglucosamine is variable. Urea, esculin and gelatin are not hydrolyzed. Presence of nitrate reductase is variable. DNase is absent. Esterase and naphthol-AS-BI-phosphohydrolase activities are detected, and esterase lipase, leucine arylamidase, valine arylamidase, cystine arylamidase, acid phosphatase, and alkaline phosphatase production is variable. No activity is detected for α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, α-fucosidase, lipase (C14), trypsin, or α-chymotrypsin. The following substrates are utilized in 2 or 4 d: D-glucose, sucrose, D-ribose, glycerol, L-malate, 2-ketogluconate, succinate, fumarate, L-aspartate, L-glutamate, L-proline, and L -serine. The following substrates are not utilized in 6 d: caprate, citrate, and DL-glycerate. The type strain has all the properties given for the species except that it also assimilates D-mannose.
Corynemycolic acids are present, and the Cellular fatty acids are consistent with those described for the genus as well as for CDC group G (Bernard et al., 1991); strains usually contain small volumes of tuberculostearic acid.
Source: a case of lepromatous leprosy in the Phillipines and a wide variety of human clinical specimens.
DNA G+C content (mol%): not available.
Type strain: Medalle X, LDC-20, ATCC 35692, CCUG 45418, CIP 107291.
Sequence accession no. (16S rRNA gene): AJ438050, X84247.
Further comments: additional GenBank accession numbers include (partial rpoB gene) AY581869.
ATCC 35529, listed as the type strain of Corynebacterium tuberculostearicum sp. nov., was deaccessioned by the ATCC and replaced with ATCC 35692 (Feurer et al., 2005).
By 16S rRNA gene sequencing, some strains previously identified as CDC group G or as “Corynebacterium pseudogenitalium”, including CIP 106714, are closely related (have >99% identity by 16S rRNA gene sequencing) to Corynebacterium tuberculostearicum (K. Bernard and G. Funke, personal communication). In 1984, “Corynebacterium tuberculostearicum” was first described as a novel species but not validly named, with the type strain being recovered from bone marrow of a patient with lepromatous leprosy in the Philippines (Brown et al., 1984). This taxon was re-evaluated, with additional clinical strains described from inguinal node, lymph node, blood culture, urethra, skin, peritoneum, urine, and a healthy urogenital tract. Also recovered from tuna and an industrial environment.
Corynebacterium tuscaniense
corrig. Riegel, Creti, Mattei, Nieri and von Hunolstein 2006b, 2025VP (Effective publication: Riegel, Creti, Mattei, Nieri and von Hunolstein 2006a, 311.)
tus.ca.ni.en'se. L. neut. adj. tuscaniense pertaining to Tuscania, Latin name of the Italian region, Tuscany, where the type strain was isolated.
Note: “Corynebacterium tuscaniae” epithet was corrected to Corynebacterium tuscaniense (Euzéby, 2006).
Gram-stain-positive rods. Colonies are whitish, circular with entire edges, nonhemolytic, grow aerobically and in the presence of a 5% CO2-enriched atmosphere. Acid produced from glucose and maltose but not from ribose, xylose, mannitol, lactose, sucrose, or glycogen. Nitrate not reduced. Produces pyrazinamidase and alkaline phosphatase but not pyrrolidonyl arylamidase, β-glucuronidase, β-galactosidase, α-glucosidase, β-N-acetyl-glucosaminidase, urease, or esterase activity. Hydrolyzes hippurate but not tyrosine, gelatin, or esculin. CAMP test and reverse CAMP test negative.
Contains mycolic acids with short chain lengths (C26–C36), but other chemotaxonomic features such as Cellular fatty acids not extant.
Source: the blood of a patient with endocarditis.
DNA G+C content (mol%): not available.
Type strain: ATCC BAA-1141, CCUG 51321, ISS-5309, JCM 15294.
Sequence accession no. (16S rRNA gene): AY677186.
Further comments: additional GenBank accession numbers include (partial rpoB gene) FJ268581.
Corynebacterium ulcerans
(ex Gilbert and Stewart 1927) Riegel, Ruimy, De Briel, Prévost, Jehl, Christen and Monteil 1995b, 619VP (Effective publication: Riegel, Ruimy, De Briel, Prévost, Jehl, Christen and Monteil 1995c, 275.) (“Corynebacterium ulcerans” Gilbert and Stewart 1927)
ul'ce.rans. L. part. adj. ulcerans making sore, causing to ulcerate.
Gram-stain-positive pleomorphic rods arranged in palisades or V-shaped forms. May contain metachromatic granules. Facultatively anaerobic. Colonies (1–2 mm in diameter) on 5% sheep blood agar are gray-white, exhibit a light hemolysis, and may have a dry and waxy consistency. Circular, slightly convex with an entire margin. Nitrate is not reduced to nitrite. Acid is produced from D-glucose, glycogen, maltose, and fructose but not from lactose, sucrose, D-xylose, and trehalose. Some strains produce acid from mannitol. Starch, DNA, and urea are degraded but tyrosine is not. Gelatin is hydrolyzed at 25°C and at 37°C by some strains. Esculin is not hydrolyzed. Methyl red is positive, and acetoin is not produced. Some strains grow in 6.5% NaCl. Hippurate is hydrolyzed. Alkaline phosphatase and α-glucosidase are produced, but pyrazinamidase, pyrrolidonyl arylamidase, β-glucuronidase, β-galactosidase, N-acetyl-β-glucosaminidase are not. As sole carbon sources, cells utilize glucose, arabinose, mannose, maltose and malate, with utilization of mannitol and gluconate occurring variably. Acetate, lactate, propionate, caprate, adipate, and phenylacetate are not ulitilzed. Fermentation of glycogen and starch plus urease is unique among species described here. The type strain is as described for the species but ferments mannitol and not gluconate.
Cell wall contains meso-DAP and the sugars arabinose and galactose. Short chain length mycolic acids (C26–C36) are present. Cellular fatty acids are consistent with those described for the genus, including characteristically (for Corynebacterium diphtheriae, Corynebacterium pseudotuberculosis, and Corynebacterium ulcerans), a large volume of a C16:1 isomer (Bernard et al., 1991). Propionic acid is produced as a metabolic product (Bernard et al., 2002).
Source: animals and humans. Infection of farm animals or their milk can result in the transmission of infection to humans. Infections from companion pets have been documented to be transferred to and cause disease in humans (De Zoysa et al., 2005b; Hogg et al., 2009).
DNA G+C content (mol%): 53 (Tm).
Type strain: ATCC 51799, CCUG 2708, CIP 106504, DSM 46325, JCM 10387, NCTC 7910.
Sequence accession no. (16S rRNA gene): X84256.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492271. Corynebacterium pseudotuberculosis and Corynebacterium ulcerans are the only Corynebacterium species to produce phospholipase D (PLD), with expression of the pld gene product being detected by inhibition of the CAMP reaction (Barksdale et al., 1981) or by PCR detection of the gene (Pacheco et al., 2007). Like Corynebacterium diphtheriae and Corynebacterium pseudotuberculosis, Corynebacterium ulcerans produces cystinase, as demonstrated by production of brown colonies and haloes when grown on modified Tinsdale medium, and fails to produce pyrazinamidase (PYRa). By 16S rRNA gene sequencing, this species is most closely related to (>99% identity with) Corynebacterium pseudotuberculosis and to (>97% identity with) Corynebacterium diphtheriae but can be readily distinguished when rpoB gene sequences are compared (Khamis et al., 2004). Corynebacterium ulcerans is able to harbor the bacteriophage which bears diphtheria toxin gene and also express the toxin, causing diphtherial like disease. If recovered from pseudomembranous materials, patient must be treated as for diphtheria.
Corynebacterium ulceribovis
Yassin 2009, 36VP
ul.ce.ri.bo'vis. L. n. ulcus, −eris ulcer; L. n. bos, bovis a cow a bull; N.L. gen. n. ulceribovis of an ulcer of a cow.
Gram-stain-positive rods. Colonies are creamy, circular, smooth, entire, and 2–4 mm in diameter after 48 h incubation at 37°C on Columbia agar supplemented with 5% sheep blood. Facultatively anaerobic. Colonies are non-hemolytic. Hippurate and Tween 80 are hydrolyzed, but esculin, gelatin, and urea are not. Activity is detected for acid phosphatase, alkaline phosphatase, esterase lipase, leucine arylamidase, pyrazinamidase, and naphthol-AS-BI-phosphohydrolase but not for arginine dihydrolase, chymotrypsin, cysteine arylamidase, esterase, α-fucosidase, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, lipase, α-mannosidase, pyrrolidonyl arylamidase, trypsin, or valine arylamidase. Nitrate is not reduced. Acetoin production is positive.
Corynemycolic acids are present. Cellular fatty acids are consistent with those described for the genus, but tuberculostearic acid is not detected.
Source: the skin of the udder of a cow with a profound ulceration in Schleswig Holstein, Germany.
DNA G+C content (mol%): not available.
Type strain: IMMIB L-1395, CCUG 55727, DSM 45146.
Sequence accession no. (16S rRNA gene): AM922112.
Corynebacterium urealyticum
Pitcher, Soto, Soriano and Valero-Guillén 1992, 180VP
u.re.a.ly'ti.cum. N.L. fem. n. urea urea; N.L. neut. adj. lyticum (from Gr. neut. adj. lutikon) able to loosen, able to dissolve; N.L. neut. adj. urealyticum urea dissolving.
Gram-stain-positive rods which may become coccoidal after prolonged culture, are 0.5–1 µm arranged in palisades and V shapes, with no tendency to branch. Bacteria grow on blood agar as pinpoint colonies after 48 h of incubation at 25, 37, and 42°C. Colonies are whitish, opaque, smooth, convex, and nonhemolytic. Respiratory metabolism, i.e. does not grow on blood agar incubated anaerobically for 48 h or on MacConkey agar. Nonreactive with usual sugars, i.e. acid is not produced from glucose, sucrose, maltose, mannitol, xylose, ribose, L-arabinose, sorbitol, lactose, trehalose, inulin, raffinose, starch, and glycogen. Does not hydrolyze gelatin or esculin. Nitrate is not reduced. Does not degrade DNA. Possesses strong urease activity. Hydrolyzes Tween 80. Lipophilic, i.e. growth is stimulated by Tween 80. Some strains hydrolyze hippurate or give a positive Voges–Proskauer reaction.
Cell wall contains meso-DAP and the sugars arabinose and galactose. Cellular fatty acids are consistent with those described for the genus and 10-methyloctadecanoic (tuberculostearic) acid is detected. Mycolic acids (C26–C36) are present. Menaquinone MK-9(H2) is found. Contains diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol, and phosphatidylinositol mannosides. Propionic acid is not produced by anaerobic metabolism of glucose.
Source: urine, particularly in humans with lithiasis and alkaline-encrusted cystitis as complications, but also recovered from other normally sterile sites and human skin. Occasionally recovered from animals with urosepsis (Bailiff et al., 2005).
DNA G+C content (mol%): 61–62 (Tm; Pitcher et al., 1992), 65–66 (HPLC, Riegel et al., 1992), and 64.2 (complete genome sequence, Tauch et al., 2008b).
Type strain: ATCC 43042, CCUG 18158, CIP 103524, DSM 7109, JCM 10395, LMG 19041, NCTC 12011.
Sequence accession no. (16S rRNA gene): X81913, X84439.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492275. Corynebacterium urealyticum Pitcher et al. 1992 was previously known as CDC coryneform group D-2 (Riegel et al., 1992). The complete genome of Corynebacterium urealyticum strain DSM 7109T, with 2,369,219 bp has been deposited in GenBank under accession no. NC_010545 (Tauch et al., 2008b). Clinical features with respect to data inferred from complete genomic analyses have been reviewed (Soriano and Tauch, 2008). Strains can be multidrug resistant (Fernandez-Roblas et al., 2009; Philippon and Bimet, 1990) and nosocomially acquired (Famularo et al., 2008).
Corynebacterium ureicelerivorans
Yassin 2007, 1202VP
u.re.i.ce.le.ri.vo'rans. N.L. fem. n. urea urea; L. adj. celer, −eris fast; L. part. adj. vorans devouring; N.L. part. adj. ureicelerivorans fast urea-devouring, referring to the rapid utilization of urea.
Gram-stain-positive rods. Colonies are creamy, circular, dry, and approximately 0.1–0.3 mm in diameter on Columbia blood agar after 48 h incubation at 37°C. Colonies are nonhemolytic. Facultatively anaerobic. Lipophilic. Urea rapidly positive (reaction in approximately 60 sec). Hippurate hydrolyzed, but esculin and gelatin are not. Nitrate not reduced. Acid produced from glucose. Weak acid production is observed from ribose and D-xylose in the API Coryne and API 20 Strep systems after 3 d. Acid is not produced from L-arabinose, glycerol, glycogen, inulin, lactose, maltose, mannitol, sorbitol, sucrose, trehalose, or D-raffinose. Alkaline and acid phosphatases, ester lipase, naphthol- AS-BI-phosphohydrolase, leucine arylamidase, pyrrolidonyl arylamidase, and pyrazinamidase are detected, but arginine dihydrolase, esterase, α-glucosidase, β-glucosidase, α-galactosidase, β-galactosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, α-mannosidase, α-fucosidase, chymotrypsin, trypsin, valine arylamidase, cystine arylamidase, and leucine arylamidase, are not. Acetoin produced.
Mycolic acids are present. Cellular fatty acids are consistent with those described for the genus, and tuberculostearic acid is present.
Source: blood culture of patient with septicemia (Yassin, 2007), but also recovered from other normally sterile body fluids (Fernandez-Natal et al., 2009).
DNA G+C content (mol%): not available.
Type strain: IMMID RIV-2301, CCUG 53377, DSM 45051, JCM 15295.
Sequence accession no. (16S rRNA gene): AM397636.
Corynebacterium variabile
corrig. (Müller 1961) Collins 1987a, 287VP (Arthrobacter variabilis Müller 1961, 524) Note: original spelling of the specific epithet, Corynebacterium variabilis (sic), was later corrected to Corynebacterium variabile.
va.ri.a'bi.le. L. neut. adj. variabile changeable, variable.
Gram-stain-positive rod-shaped cells (0.8–1.1 × 1.4–3.5 µm), irregularly shaped (club-shaped or tapered), and occurring singly, in pairs with typical V forms, or clumps; ovoid forms occur in older cultures. Colonies are small (∼2–4 mm), circular (sometimes irregular), convex, and gray-white (occasionally slight pink) with a dry appearance. Optimum temperature is 25–30°C. Grows in 7% NaCl. Strictly aerobic. Acetate, propionate, capronate, 4-aminobutyrate, caprylate, succinate, DL-malate, levulinate, and some other compounds may be used as sole carbon sources. Xanthine, tyrosine, and starch are not hydrolyzed.
Cell wall contains meso-DAP, with the glycan moiety containing only acetyl residues. The cell wall also contains an arabinogalactan polymer. Short-chain mycolic acids (C30–C36 carbon atoms) are present. Cellular fatty acids are consistent with those described for the genus, including high levels of tuberculostearic acid. The major menaquinones are MK-9(H2) and MK-8(H2).
Source: the environment and the surface of soft cheese (Gelsomino et al., 2005).
DNA G+C content (mol%): 65 (Tm).
Type strain: ATCC 15753, CCUG 45246, CIP 102112, DSM 20132, HAMBI 1872, JCM 2154, NBRC 15286, NCIMB 9455, NRRL B-4201, VKM Ac-1122.
Sequence accession no. (16S rRNA gene): AJ222815.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492272. Arthrobacter variabilis Müller 1961AL and Caseobacter polymorphus Crombach 1978AL are both considered as later synonyms of Corynebacterium variabile (Collins 1987a, 1989). Corynebacterium mooreparkense (Brennan et al., 2001) is also considered as a later heterotypic synonym of Corynebacterium variabile corrig. (Müller 1961) Collins 1987a, 287 (Gelsomino, Vancanneyt, Snauwaert, Vandemeulebroecke, Hoste, Cogan and Swings 2005, 1129).
Corynebacterium vitaeruminis
corrig. (Bechdel et al. 1928) Lanéelle, Asselineau, Welby, Norgard, Imaeda, Pollice and Barksdale 1980, 544VP [“Flavobacterium vitarumen” Bechdel, Honeywell, Dutcher and Knutsen 1928, 234; Brevibacterium vitarumen (Bechdel et al. 1928) Breed, Murray and Smith 1957, 495]
vi.ta.e.ru.mi'nis. L. n. vitae life; L. n. rumen -inis throat, gullet, rumen; N.L. gen. n. vitaeruminis of rumen life.
Note: spelling of the epithet, vitarumen (sic), was corrected by Trüper and De'Clari 1997, 908.
(Some information from this description is based on that of Collins and Cummins, 1986.)
Pleomorphic Gram-stain-positive rod-shaped bacteria wth moderately tapered ends. May contain metachromatic granules when grown on phosphate rich media. Lemon yellow colonies on Loeffler slants with raised, smooth, butyrous yellow colonies on chocolate agar, and black colonies with chocolate agar containing 0.03% tellurite. Facultatively anaerobic. Positive for glucose, fructose, galactose, mannose, maltose, sucrose, salicin, and trehalose. Negative for glycerol, inositol, mannitol, lactose, and starch. Esculin hydrolyzed, nitrate is reduced to nitrite, acetoin and urease are produced, and methyl red is positive. Pyrazinamidase positive, but phosphatase negative. Hippurate and gelatin are not hydrolyzed.
Cell wall contains meso-DAP and the sugars arabionse and galactose. Short-chain corynemycolates and menaquinone MK-8(H2) are present.
Source: the rumen of a cow.
DNA G+C content (mol%): 64.8 (Tm).
Type strain: ATCC 10234, CCUG 28792, CIP 82.7, DSM 20294, JCM 1323, NBRC 12143, NCIMB 9291, VKM B-1211.
Sequence accession no. (16S rRNA gene): AY438066.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492273.
Corynebacterium xerosis
(Lehmann and Neumann 1896) Lehmann and Neumann 1899, 385AL (Bacillus xerosis Lehmann and Neumann 1896, 361)
xe.ro'sis. N.L. gen. n. xerosis (from Gr. adj. xeros dry) of xerosis.
Note: description below is based on ATCC 373T, as some commercially available reference strains were shown to be misidentified after contemporary characterization methods were applied, with some being identifiable as Corynebacterium striatum (Coyle et al., 1993b) or Corynebacterium amycolatum (Funke et al., 1996a; Wauters et al., 1996).
Gram-stain-positive rods with irregular staining, barred rods with occasional granules and club forms. Colonies are small, circular ∼0.2–1.0 mm in diameter after growth on blood agar at 24 h. Colonies dryish and slightly yellow colored (Coyle et al., 1993b). In broth, forms a large pellicle over a clear broth, with settled clumps observed. Non-hemolytic. Not lipophilic. Facultatively anaerobic. Grows anaerobically. CAMP reaction negative. Nitrate either reduced or considered as variable (Funke and Bernard, 2007). Glucose, maltose, and sucrose fermented, but xylose and mannitol are not (Coyle et al., 1993b; Funke and Bernard, 2007). Also described as usually being negative for arabinose, rhamnose, lactose, trehalose, raffinose, dextrin, and starch, but positive for salicin (Collins and Cummins, 1986). ATCC 373T has been described as containing cell-wall sugars of arabinose, galactose, and glucose with smaller amounts of rhamnose. Corynemycolates are present. Cellular fatty acids consistent with those described for the genus are present, but tuberculostearic acid is not detected (Bernard et al., 1991). Propionic acid is not detected as a fermentation product (Funke et al., 1996a).
Source: the ear discharge of a child.
DNA G+C content (mol%): 67.3 (method unknown).
Type strain: ATCC 373, CCUG 27544, CIP 100653, DSM 20743, JCM 1971, NBRC 16721, NCTC 11861.
Sequence accession no. (16S rRNA gene): X81914, X84446.
Further comments: additional GenBank accession numbers include (complete rpoB gene) AY492233. Once described as a relatively common human pathogen, Corynebacterium xerosis infections are now considered to occur only very rarely, and diseases attributed to this agent should be considered as definitive only if polyphasic, including genetic identification methods, are used. In particular, older publications implicating Corynebacterium xerosis as a pathogen, especially those where microbiological descriptions were based solely on phenotypic testing, may be actually describing disease caused by Corynebacterium amycolatum Funke et al. 1996a or other Corynebacterium species. Reference strains other than ATCC 373T designated Corynebacterium xerosis must be carefully evaluated prior to use in research. ATCC 7711, with a DNA G+C content of 68.5 mol% (Collins and Cummins, 1986), has been found to be closely related to ATCC 373T by 16SrRNA and partial rpoB gene sequence analyses, but NCTC 7243 is closely related to (>99% identity with) Corynebacterium amycolatum (K. Bernard, personal communication). Corynebacterium xerosis cannot be easily discerned from Corynebacterium freneyi and Corynebacterium hansenii by 16S rRNA, rpoB, or 16S–23S spacer region gene sequence analyses; these are best separated by DNA–DNA hybridization or phenotypic means (Renaud et al., 2007).



