Chronic lymphocytic leukemia B cells are endowed with the capacity to attract CD4+, CD40L+ T cells by producing CCL22
Abstract
The natural history of B-chronic lymphocytic leukemia (CLL) is not entirely explained by intrinsic defects of the neoplastic cell, but is also favored by microenvironmental signals. As CLL cells retain the capacity to respond to CD40 ligand (CD40L) and as CD4+ T cells are always present in involved tissues, we asked whether malignant CLL cells might produce T cell-attracting chemokines. We studied the chemokine expression of CD19+/CD5+ malignant B cells from peripheral blood (PB), lymph nodes (LN) or bone marrow (BM) of 32 patients and found a major difference. LN- and BM-, but not PB-derived cells, expressed a readily detectable reverse transcription-PCR band for CCL22 and one for CCL17 of variable intensity. CD40 ligation of PB cells induced the mRNA expression of both CCL22 and CCL17. CCL22 was also released in the culture supernatants. These supernatants induced the migration of activated CD4+, CD40L+ T cells expressing the CCL22 receptor, CCR4. T cell migration was abrogated by anti-CCL22 antibodies. Immunohistochemistry and cytofluorography studies revealed that a proportion of CD4+ T cells in CLL LN and BM expressed CD40L. Our data demonstrate that malignant CLL cells chemo-attract CD4+ T cells that in turn induce a strong chemokine production by the leukemic clone, suggesting a vicious circle, leading to the progressive accumulation of the neoplastic cells.
Abbreviations:
-
- CLL:
-
Chronic lymphocytic leukemia
-
- CR:
-
Chemokine receptors
-
- PB:
-
Peripheral blood
-
- FACS:
-
Fluorescence-activated cell sorting
1 Introduction
Chemokines (chemoattractant cytokines) are a rapidly expanding group of small (8–14 kDa) secreted molecules, recently classified according to their structure 1, that regulate the migration and homing of leukocytes during immune and inflammatory reactions. At least 16 different G protein-coupled seven transmembrane domain molecules, the chemokine receptors (CR) 2, 3, mediate the functional activity of chemokines. CR are divided into CC or CXC receptors depending on which chemokine they bind.
T cells are the best-studied leukocytes in terms of CR expression and response to chemokines. The trafficking of different T cells subpopulations is dependent upon their specific CR expression, which in turn is tightly regulated by the cell's antigenic experience, state of activation and functional polarization. CCR4, the receptor for the chemokines CCL22/MDC 4–6 and CCL17/TARC 7, 8, is constitutively expressed by CD4+ Th2 cells 9–12 and is up-regulated on both Th1 and Th2 cells upon TCR and CD28 engagement 13. Therefore, CCR4 exerts a central role in physiological immune responses by attracting activated CD4+ T cells to sites of antigenic challenge and/or to specialized microenvironments within lymphoid tissues 13.
Chemokines appear to play an important role in tumor growth and expansion through autocrine or paracrine amplification mechanisms 14. Tumor cells themselves and/or surrounding non-tumor cells indeed produce chemokines which favor the tissue accumulation of malignant cells 15, 16. Also, tumor cell transduction of chemokine genes, either alone or in combination with other immunoregulatory genes, has been shown to significantly improve the generation of anti-tumor immunity in animal models 17, 18. We have asked whether primary tumor cells may produce chemokines able to attract by-stander cells thereby influencing the organization of the microenvironment where tumor cells themselves grow. This possibility is intriguing as it implies that primary tumor cells actively participate in the generation of the tumor microenvironment by recruiting cells that in turn might favor the tumor cell expansion and/or survival.
To this end, an interesting model is provided by B cell chronic lymphocytic leukemia (CLL), a malignancy characterized by the progressive accumulation of resting CD5+ B lymphocytes, which are resistant to apoptosis 19. Such a resistance can not be explained only by intrinsic defects of the neoplastic cell, but seems to be largely due to the fundamental (thoughnot sufficient) role of by-stander, non-tumor cells and the factors they produce 20. There is a substantial evidence that malignant CLL cells retain the capacity to respond to microenvironmental signals, such as those delivered by CD40 ligand (CD40L)+ Th cells. CD40 stimulation rescues CLL cells from apoptosis and induces their proliferation 21, 22. Along this line, we have recently reported that CD40 cross-linking, can improve the survival of CLL cells, by up-regulating the expression of the anti-apoptotic gene survivin 22. Further, the presence of high numbers of CD4+ T cells is evident in CLL pseudo-germinal centers (GC), the proliferating cell reservoir located in invaded bone marrow (BM) and lymph nodes (LN) 23.
In this work we show that CLL cells purified from involved LN and BM, but not from peripheral blood (PB), constitutively express mRNA for the T cell attracting chemokines CCL17 and CCL22. In asearch of signals delivered by the microenvironment that could account for such a difference, we demonstrate that CD40-cross-linking of PB CLL cells induces the expression of both chemokines at RNAlevel. Of the two, only CCL22 is released and is capable of attracting activated CD4+ CD40L+ T cells. Also, we provide evidence that a proportion of CD4+ T cells interspersed within the proliferating CLL cell clusters in the LN are CD40L+, suggesting the in vivo availability of this signal to leukemic cells. Taken together these findings indicate that the stimulation of malignant cells via a physiological signal present in the tumor microenvironment endows CLL cells with the chemoattracting capacity for CD4+ T cells, which in turn can deliver survival signals to tumor cells.
2 Results
2.1 Chemokine expression by CLL cells from PB, BM and LN
As several evidences indicate that CLL cells retain the capacity to respond to microenvironmental signals delivered by activated T lymphocytes and as T lymphocytes are present in CLL LN and BM, we hypothesized that CLL cells might produce T cell chemoattractants. Therefore, our first step was to analyze neoplastic B lymphocytes purified from PB of 24 cases (Pt. 1–24, Table 1), BM of 4 other patients (Pt. 25–28, Table 1) and LN of 4 further cases (Pt. 29–32, Table 1).
All cases from LN, BM and 7 cases from PB were purified by cell sorting. Malignant B lymphocytes from the remaining PB were not purified, due to the high percentage of leukemic cells present in the preparations (>95%).
As shown in Table 2, only few chemokines could be detected by RT-PCR in LN-, BM- and PB-derived primary leukemia cells and at variable intensity.
CCL5 was detected in all samples tested regardless of their origin (Table 2 and Fig. 1A). CCL3 was absent in 4 PB cases (Table 2), and expressed, at variable levels, in all other samples tested (Fig. 1A). CXCL8 was absent in 12 samples (2 LN, 1 BM and 9 PB) (Table 2) anddetectable in the remaining tested cases (Fig. 1A). Interestingly, CLL cells obtained from LN and BM expressed a readily detectable band for CCL22 (in 4/4 LN cases and 2/4 BM cases) and a band of variable intensity for CCL17 (in 2/4 LN cases and 0/4 BM cases) (Fig. 1B). In contrast, both chemokines were absent in PB-derived cells (0/24 cases) (Fig. 1B).
|
Patient |
R/NRa) |
Se |
Age |
Stage |
Isotype/light chain |
CD38 |
Sample |
|---|---|---|---|---|---|---|---|
|
1 |
R |
M |
65 |
0 |
IgM/κ |
1 % |
PB |
|
2 |
R |
F |
76 |
0 |
IgG/λ |
6 % |
PB |
|
3 |
R |
F |
80 |
0 |
IgM-D/λ |
92 % |
PB |
|
4 |
R |
M |
67 |
0 |
IgM-D/κ |
85 % |
PB |
|
5 |
R |
M |
67 |
0 |
IgM-D/κ |
21 % |
PB |
|
6 |
R |
M |
73 |
II |
IgM-D/κ |
39 % |
PB |
|
7 |
R |
M |
68 |
II |
IgM-D/κ |
25 % |
PB |
|
8 |
R |
M |
51 |
II |
IgM-D/κ |
11 % |
PB |
|
9 |
R |
F |
79 |
III |
IgM-D/λ |
54 % |
PB |
|
10 |
R |
F |
80 |
III |
IgM-D/λ |
66 % |
PB |
|
11 |
R |
F |
68 |
III |
IgM-D/λ |
9 % |
PB |
|
12 |
R |
M |
81 |
IV |
IgM-D/λ |
33 % |
PB |
|
13 |
R |
M |
77 |
IV |
IgM-D/κ |
65 % |
PB |
|
14 |
R |
M |
72 |
IV |
IgM-D/κ |
6 % |
PB |
|
15 |
NR |
F |
68 |
0 |
IgM-D/κ |
16 % |
PB |
|
16 |
NR |
F |
64 |
0 |
IgM-D/λ |
10 % |
PB |
|
17 |
NR |
F |
75 |
0 |
IgM-D/κ |
66 % |
PB |
|
18 |
NR |
M |
72 |
0 |
IgM-D/λ |
30 % |
PB |
|
19 |
NR |
M |
85 |
II |
IgM-D/κ |
62 % |
PB |
|
20 |
NR |
M |
52 |
IV |
IgM-D/κ |
1 % |
PB |
|
21 |
NR |
F |
67 |
IV |
IgG/κ |
6 % |
PB |
|
22 |
ND |
F |
61 |
0 |
IgM-D/λ |
18 % |
PB |
|
23 |
ND |
M |
73 |
0 |
IgM/λ |
2 % |
PB |
|
24 |
ND |
F |
66 |
II |
IgM-D/κ |
39 % |
PB |
|
25 |
ND |
F |
64 |
I |
IgM-D/κ |
16 % |
BM |
|
26 |
ND |
M |
67 |
II |
IgM-D/κ |
82 % |
BM |
|
27 |
ND |
M |
70 |
III |
IgM-D/λ |
9 % |
BM |
|
28 |
ND |
F |
53 |
IV |
IgM-D/κ |
54 % |
BM |
|
29 |
ND |
M |
60 |
I |
IgM-D/κ |
– |
LN |
|
30 |
ND |
M |
65 |
II |
IgM-D/λ |
– |
LN |
|
31 |
ND |
M |
51 |
II |
IgM/λ |
62 % |
LN |
|
32 |
ND |
M |
70 |
III |
IgM-D/λ |
12 % |
LN |
- a) R: CD40L-responder; NR: CD40L non-responder; ND not done.
|
|
Unstimulated CLL cells |
||
|---|---|---|---|
|
C-C Chemokine |
LN |
BM |
PB |
|
CCL22/MDC |
4/4 |
2/4 |
0/24 |
|
CCL17/TARC |
2/4 |
0/4 |
0/24 |
|
CCL3/MIP-1α |
4/4 |
2/2 |
16/20 |
|
CCL5/RANTES |
4/4 |
2/2 |
20/20 |
|
CCL20/LARC |
0/4 |
0/2 |
0/12 |
|
CCL21/SLC |
0/4 |
0/2 |
0/12 |
|
CCL19/ELC |
0/4 |
0/2 |
0/12 |
|
CCL1/I-309 |
0/4 |
0/2 |
0/12 |
|
CCL25/TECK |
0/4 |
0/2 |
0/12 |
|
CCL18/PARC |
0/4 |
0/2 |
0/12 |
|
C-X-C Chemokines |
|||
|
CXCL8/IL-8 |
2/4 |
1/2 |
15/24 |
|
CXCL10/IP-10 |
0/4 |
0/2 |
0/12 |
|
CXCL9/MIG |
0/4 |
0/2 |
0/12 |
|
CXCL12/SDF-1α |
0/4 |
0/2 |
0/12 |
|
CXCL12/SDF-1β |
0/4 |
0/2 |
0/12 |
|
CXCL13/BCA-1 |
0/4 |
0/2 |
0/12 |
|
C-X3-C Chemokine |
|||
|
CXC3CL1/Fractalkine |
0/4 |
0/2 |
0/12 |
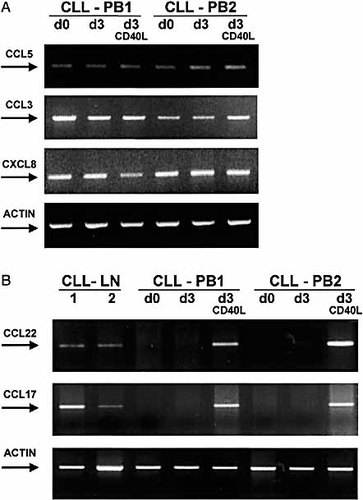
(A) CCL5, CCL3 and CXCL8 mRNA expression in PB-derived CLL cells. CLL cells from two representative PB (CLL-PB1 d0 and CLL-PB2 d0) express CCL5, CCL3, and CXCL8 mRNA. The expression levels are not modified after 3 days of in vitro culture in the absence or the presence of sCD40L (d3 and d3 CD40L). (B) CCL22 and CCL17 mRNA expression in LN- and PB-derived CLL cells. CLL cells from two representative LN (CLL-LN 1 and 2) express CCL22 and CCL17 mRNA, while CLL cells derived from two representative PB (CLL-PB1 d0 and CLL-PB2 d0) do not. CD40 cross-linking of PB-derived cells induces the expression of mRNA for CCL22 and CCL17, after 3 days of culture (d3 CD40L). Leukemia cells were also cultured in the absence of recombinant sCD40L, for 3 days (d3), without any detectable induction of both chemokines. LN and PB-derived CLL cells were FACS-sorted obtaining >99% purity.
2.2 CD40 cross-linking of leukemia cells induces RNA expression of the chemokines CCL17 and CCL22
These results suggested that signals available in the LN microenvironment might account for the different behavior of tissue-derived as compared to PB-derived malignant B cells. We focussed our attention on the CD40/CD40L interaction, which is the hallmark of T/B cell interactions in secondary lymphoid tissues. Fluorescent-activated cell sorting (FACS)-purified CLL cells from 7 patients' PB were cultured either in medium alone or in the presence of recombinant sCD40L for up to 3 days. CD40 ligation induced mRNA expression of both CCL22 and CCL17 (Fig. 1B), without altering the level of expression of either CCL3, or CCL5 or CXCL8 (Fig. 1A). Both CCL22 and CCL17 started being expressed already after 24 h of culture (data not shown) and maintained the same level of expression throughout the 3 days of culture (Fig. 1B). These results were then confirmed in 14 further CLL cases, whose malignant cells were not purified, due to the high percentage of leukemic cells present in the preparations (>95%).
2.3 CD40 cross-linking induces the secretion of CCL22 chemokine
Starting from these evidences, we next sought to determine whether CCL22 and CCL17 chemokines were actually secreted by leukemic cells and whether their production might be modified by cell activation. We prepared leukemic cell supernatants by culturing CLL cells at standard cell concentration (2×106 cells/ml), in the presence or the absence of sCD40L. We collected the culture supernantants after three days of culture and measured by ELISA the presence of chemokines. CCL3, CCL5, CCL17 and CXCL8 were measured in 12 cases and CCL22 in 21 cases. Leukemic cells produced low levels (up to 0.36 ng/ml) of CCL3 in 8/12 cases and low levels (up to 3.16 ng/ml) of CXCL8 in 6/12 cases. CCL5 protein was detectable in all 12 supernatants tested, though at very variable levels, ranging between 0.07 to 36 ng/ml. The production of all these chemokines was not dependent on the culture condition i.e. no significant modifications occurred after CD40 activation.
Supernatants from unstimulated CLL cells were negative for both CCL17 and CCL22 and upon CD40-stimulation no CCL17 could be detected in the cell supernatants (data not shown). In contrast, CD40-stimulated malignant cells produced levels of CCL22 as high as 28.8 ng/ml (Fig. 2).
We investigated whether cell density might account for the difference between CCL17 and CCL22. CLL cells were cultured at high-density concentration (10×106 cells/ml) but still no CCL17 was observed. On the contrary, the supernatant levels of CCL22 paralleled cell density (data not shown). To confirm that CCL22 was actually secreted by tumor cells we tested supernatants obtained from purified (>99% purity) malignant CLL cells of seven patients. Sorted cells produced levels of CCL22 comparable to those of the unsorted samples.
Not all patients produced CCL22 after CD40-stimulation. Two categories of patients were observed: CD40L-responders and CD40L-nonresponders. In 14/21 (67%) patients studied (patients 1–14, Table 1) CD40L stimulation induced CCL22 release in the supernatants. These patients were considered CD40L-responders. The remaining 7/21 cases (33%; Table 1: patients 15–21) were considered CD40L-nonresponders as CCL22 secretion remained unchanged after stimulation. Twenty patients of this series have been previously analyzed for the capacity to express Survivin mRNA after CD40 stimulation 22. In 17 out of these 20 patients, there was correspondence in the outcome of the response.
The ability of responding to CD40 stimulation did not correlate with Rai stage of the disease nor with CD38 expression, though the limited number of patients studied prevented any statistically significant conclusion.
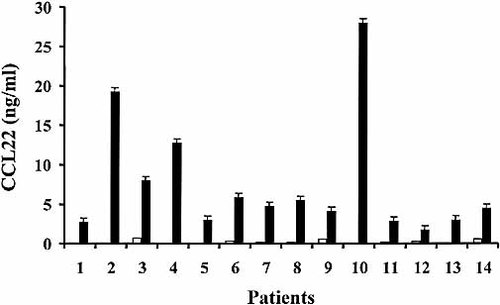
CD40 cross-linking of CLL cells induces the secretion of CCL22. Leukemia cells were cultured in control medium (white bars) or in the presence of recombinant sCD40L (black bars) for 3 days and supernatants collected. Fourteen CD40L-responders are shown. The limit of detection of the ELISA was 0.0625 ng/ml.
2.4 Secreted CCL22 is functional and induces the migration of activated CCR4+, CD4+ and CD40L+ T cells
To determine whether CCL22 produced by CD40-stimulated CLL cells was functional, supernatants were tested for their capacity to induce the migration of activated CD4+ T cells 5, 11. To this end T cells were obtained from PBMC sorted for the expression of CCR4 and activated in the presence of irradiated feeder and IL-2, as described in the Sect. 4. As a result, these cells were CD4+ and uniformly expressed CD40L within 24 h from activation (Fig. 3). We studied these cells for the expression of several known chemokine receptors and in particular for the expression of CCR1, CCR3, CCR5, receptors for CCL3 and CCL5; CXCR1 and 2, receptors for CXCL8. T cell lines were negative for all these receptors. In contrast they uniformly expressed high levels of CCR4, the receptor for CCL22 (and CCL17) (Fig. 3).
Accordingly, recombinant CCL22, used as control, efficiently promoted the migration of recently activated T cells (Fig. 4A).
As shown in Fig. 4B, supernatants from CD40-stimulated CLL cells, obtained from six different patients, induced migration of activated T cells in a proportion significantly higher than that induced by control culture supernatants. As expected, the supernatant-induced migration was lower than that induced by recombinant CCL22 since the total amount of CCL22 present in the supernatants was lower than the optimal dose of chemokine necessary to induce optimal migration.
To demonstrate that the T cell chemotactic activity of the supernatants was specifically mediated by the CCL22 present in our supernatants, migration experiments were repeated using neutralizing antibodies. The use of anti-CCL22 antibody resulted in complete neutralization of the T cell migration to background levels (Fig. 4B). These results demonstrate that CCL22 is the only mediator of the T cell chemotactic activity present in the supernatants from CD40-stimulated CLL cells.
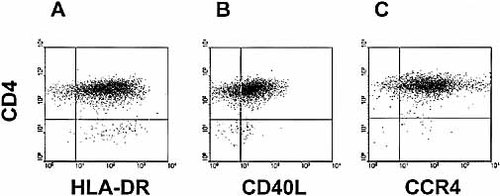
CD4+ T cells express CD40L and CCR4. FACS analyses of one representative sample of T cells. CD4+ T cells are activated, as shown by the expression of HLA-DR (A), uniformly express CD40L (B) and CCR4 (C). CD4 expression is depicted on the y-axis and the fluorescence intensity of the other markers is shown on the x-axis.
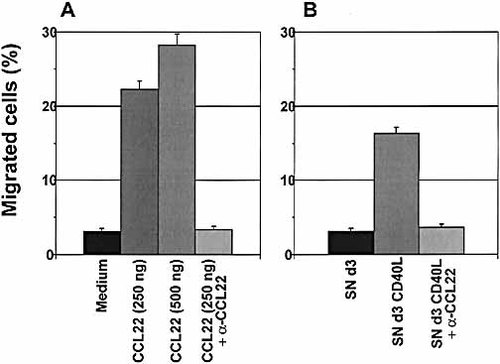
Supernatants from CD40-stimulated CLL cells induce migration of activated CD4+ T cells. (A) Recombinant CCL22 was used, as control, at 250 and 500 ng/ml and induced migration of activated T cells. Migration was blocked by adding neutralizing anti-CCL22 antibody (10 μg/ml) (CCL22 + αCCL22). (B) Supernatant from unstimulated CLL cells, cultured for 3 days in medium only (SN d3), was unable to induce migration of T cells, as compared to the control (medium in A). In contrast, supernatant from CLL cells after 3 days of culture in the presence of sCD40L (SN d3 CD40L) induced specific migration of T cells, inhibited by the addition of anti-CCL22 antibody (SN d3 CD40L + αCCL22). Data show one out of six patients studied, for both unstimulated and stimulated cells. Data are expressed as percentage of migrated T cells.
2.5 CD4+ CD40L+ T cells present in CLL LN and BM are interspersed within pseudo-follicles
Finally, as in vitro data were ascribing a role to CD40 ligation in inducing chemokine regulation in CLL cells, we evaluated whether this signal (i.e. CD40L+ T cells) was present in tissue biopsies. In BM and LN samples studied by cytofluorography analysis CD40L+ cells were clearly detectable among the CD3+ CD4+ T cells, and their proportion ranged between 5 and 30% of them. No CD19+ CD5+ B cells expressed CD40L in all the samples analyzed.
We next investigated LN and BM biopsies heavily infiltrated with malignant CLL cells by immunohistochemical analysis. In double-stained sections, CD4+ T lymphocytes were admixed with CD23+ malignant B cells (Fig. 5A) 24. Within pseudo-follicles, CD4+ T cells were interspersed with Ki67+ proliferating CLL cells (Fig. 5B), and close contacts were evident. As previously shown 22, CD3+ T cells are present in high numbers and are mainly localized within pseudo-follicles. The analyses of serial sections revealed CD40L expression in a proportion of lymphocytes corresponding to CD3+ T cells, preferentially clustered within pseudo-follicles (Fig. 5C).
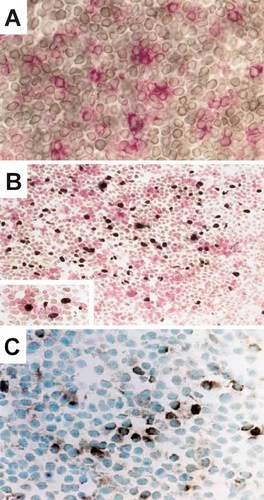
Cryostat sections from a CLL lymph node indicate that proliferating CLL cells are juxtaposed with CD4+ T cells, in pseudo-follicles. (A) Double staining of sections from the LN of a CLL patient indicates that CD4+ T lymphocytes (red immunostaining, alkaline-phosphatase anti-alkaline phosphatase; original magnification 400x) are admixed with CD23+ malignant B cells (brown immunostaining, avidin-streptavidin immunoperoxidase). (B) Within pseudo-follicles, CD4+ T cells (red immunostaining, alkaline-phosphatase anti-alkaline phosphatase) are interspersed with Ki67+ proliferating CLL cells (brown immunostaining, avidin-streptavidin peroxidase; original magnification 250x). In the insert (original magnification 400x), the close contacts between Ki67+ CLL cells and CD4+ T cells are evident. (C) CD40L expression (brown immunostaining, avidin-streptavidin immunoperoxidase; original magnification 400x) was demonstrated in a proportion of lymphocytes corresponding to CD3+ T cells in serial sections that preferentially clustered within pseudo-follicles.
3 Discussion
We have studied CLL to explore the possibility that primary tumor cells of chronic lymphoid malignancies may contribute to the development of a microenvironment suitable for their expansion and survival by producing chemokines able to attract tumor-supportive by-stander cells. The rationale of this study is threefold. First, chemokines are involved in tumor growth and expansion by means of autocrine or paracrine mechanisms 14–16. Secondly, CLL is the prototype of low-grade lymphoid malignancies, where several evidences emphasize a role of normal by-stander cells. Thirdly, CLL cells are responsive to environmental stimuli. We have recently shown that the stimulation of CD40 on the surface of CLL cells by its natural ligand triggers the expression of the anti-apoptotic gene survivin 22 and prolongs their survival.
In this work we provide evidence that CD5+ malignant cells from LN and BM have a major difference as compared to circulating cells. CLL cells purified from involved LN and BM, but notfrom PB, constitutively express mRNA for the T cell attracting chemokines CCL17 and CCL22 (Table 2 and Fig. 1B). This difference may be accounted for microenvironmental stimuli since the stimulation of CD40 on the surface of PB CLL cells induces the expression of both chemokines at RNA level (Fig. 1B). Of the two only CCL22 is released in detectable amounts (Fig. 2) and is capable of attracting activated CD4+ CD40L+ T cells (Fig. 4). As CD4+ T cells interspersed within the pseudo-follicles in the CLL LN may express CD40L+ (Fig. 5C), the in vivo availability of this signal to leukemic cells is evident.
This study further underscores that CCL22 and CCL17, originally described as specific products of myeloid cells 5, 25, are also produced by B lineage cells.To date, CCL17 and CCL22 are the only CD4+ T cell chemoattractants secreted by normal and malignant B cells, in both mice 26, 27 and humans 28, 29. Their secretion following CD40 ligation 26, 27, a critical signal in B cell biology, suggests that CCL17 and CCL22 might play an important role in the guidance of T cell-dependent B cell responses in secondary lymphoid tissues 30, 31. The sustained production of these molecules might ensure the recruitment of T cell help 9, 12, 13 which would further stimulate and drive these B cells into maturation and selection.
The paucity of chemokine production by primary CLL B cells is striking as several studies have shown that B cell receptor (BCR)-stimulated B cells up-regulate expression of chemokines, including MIP-1α and MIP-1β 32. A possible explanation lies in the fact that CLL cells have a defective signal transduction through BCR 33. It may be suggestedthat the paucity of chemokine production possibly contributes to the leukemia cells' ability to evade the host's immune system. Although other factors, such as the inadequate expression of costimulatory molecules may also account for this "immune escape", the lack of chemokine production by tumor cells may prevent the attraction of existing effector cells to the tumor site. It will be interesting to explore whether this is a more general mechanism that allow lymphoid tumors to evade tumor-specific effector T cells. In contrast, it is interesting to underscore that malignant cells maintain the capacity to produce chemokines responsible for the attraction of T cell help, which in turn can favor the malignant cell growth and survival. This seems to indicate that a remarkable feature of CLL is the selectivity in the production of "useful" versus "detrimental" chemokines.
We have shown here that CD4+, CD40L+ activated T cells are present in the involved tissues and tend to cluster around Ki67+ proliferating cells. This brings in several questions that are interesting from an immunological point of view but are also likely relevant for the natural history of CLL. Which is the activation mechanism that triggers the expression of CD40L on these T cells? Are these T cells activated following TCR engagement or in a TCR independent fashion? If the former possibility is correct, then the corollary becomes whether there is an antigen (and which antigen) that is involved in CLL genesis.
It has to be noted that in CLL up-regulation of CCL17 RNA is less relevant than that of CCL22. Also, CCL17 is not secreted, while CCL22 is. At least two possible and not mutually exclusive explanations for this different behavior may be envisaged. The first derives from the notion that the stimulation of both normal and leukemic B cell precursors 26–29up-regulates the expression of both chemokines and leads to their secretion. It is thus possible that the differential modulation of CCL22 and CCL17 may be ascribed to a different translational activity due to the cell differentiation stage and/ or to the specific tissue localization. Alternatively, it is plausible to consider that CD40L might be only one of a number of favoring microenvironmental stimuli. Other stimuli, perhaps provided by BM stromal cells and/or follicular dendritic cells, may be needed for the actual chemokine production and release. The same line of reasoning may be utilized to explain why a minority of patients is CD40L non-responders. In contrast the capacity to respond to CD40L may well reflect a difference in the natural history and the pathogenesis of the disease. Therefore, we are currently investigating in a larger series of cases whether a correlation between CD40L-responsiveness, prognostic factors and clinical outcome may exist.
In conclusion, our findings indicate that CLL cells are able to produce chemoattractant molecules (i.e. CCL22) that can selectively attract Th cells, which, through the CD40L molecule, may both provide relevant survival signals to the leukemic clone 22 and cause an increased production of T cell-attracting chemokines. This would give rise to a vicious circle, leading to the progressive accumulation of the neoplastic cells.
4 Materials and methods
4.1 Cells
Malignant B cells from 32 CLL patients were studied. Twenty-four samples were from PB, 4 were from BM and 4 from biopsied LN. Tissue samples were obtained following institutional guidelines. Diagnosis was based on standard clinical and laboratory criteria 34, 35. All patients were staged according to Rai's 36 and studied either before or at least 3 months after chemotherapy. Table 1 summarizes the main clinical data of the patients.
LN specimens were minced with a scalpel blade, reduced to single-cell suspension and passed through a fine wire mesh. All samples were enriched by density centrifugation over Lymphoprep (Nycomed Pharma AS, Oslo, Norway). When indicated, malignant B cells were purified by FACS.
CD4+ T cells were obtained from PB mononuclear cells (PBMC) of two healthy individuals.
4.2 Phenotypic analysis and cell sorting
Expression of cell surface molecules was determined by flow cytometry using standard methodology. The mAb used were: tricolor (TC)-conjugated anti-CD3, fluorescein (FITC)-conjugated anti-CD4 and anti-CD8 (Caltag, Burlingame, CA), FITC-anti-CD5 (Becton Dickinson, San Jose, CA) and phycoerythrin (PE)-anti-CD19 (Caltag), PE-anti-CD40L (DAKO A/S, Glostrup, DK); unconjugated CCR4 (IgG1), produced by us 37; PE-anti-CCR1, CCR2, CCR6, CXCR1, and CXCR2 and biotinylated-anti-CXCR5 (R&D Systems, Minneapolis, MN), PE-anti-CCR5, CXCR3 and CXCR4 (Pharmingen-Becton Dickinson,San Jose, CA). PE-conjugated goat-anti-mouse IgG1 and PE-streptavidin (Southern Biotechnologies, Birmingham, AL) were used as secondary reagents. Irrelevant isotype-matched antibodies were used as negative controls (Becton Dickinson). Samples were analyzed in a FACSCalibur flow cytometer (Becton Dickinson). At least 10,000 events were measured for each sample. For cell sorting of leukemic cells, CD5/CD19-labeled cells were concentrated, filtered, and purified using a FACStar cell sorter (Becton Dickinson). PBMC were stained with anti-CD4 and anti-CCR4 and then purified using a FACSVantage cell sorter (Becton Dickinson).
4.3 In vitro culture and CD40 stimulation of leukemic cells
Leukemic cells were cultured in RPMI medium with glutamine (Life Technologies Ltd, Paisley, Scotland) plus 10% fetal bovine serum (FBS) (Life Technologies Ltd), and gentamycin (10 μg/ml, Life Technologies Ltd) (complete RPMI-cRPMI) in the presence or the absence of 100 ng/ml soluble recombinant human CD40L plus 1 μg of enhancer, according to manufacturer's instructions (Alexis Corporation, San Diego, CA), at 37°C, 5% CO2. Malignant cells were cultured at 2×106 cells/ml and/or at 10×106 cells/ml, as indicated. Supernatants and cells were collected at day 3. Cells were then lysed in RNazol (Biotecx Laboratories, Inc., Houston, TX). Supernatants were filtered and kept frozen at –20°C.
4.4 Generation of activated CD4+ T cells
FACS-purified CCR4+ CD4+ T cells were cultured in cRPMI and activated by repetitive stimulation using irradiated allogeneic PBMC plus 1 μg/ml PHA (Murex Diagnostics) and recombinant IL-2 (20 U/ml) for 5 days.
4.5 RT-PCR
RNA was extracted from 1×106–2×106 cells using RNazol (Biotecx Laboratories, Inc., Houston, TX) following the manufacturer's instructions. cDNA was prepared at42°C using a reverse transcription mix containing Superscript II (Life Technologies Ltd). The enzyme was inactivated for 4 min at 94°C. PCR amplification of cDNA samples was carried out using the primers whose sequences are shown in Table 3.
All reactions were carried out in a Perkin-Elmer thermocycler (Perkin-Elmer, Foster City, CA), at the following conditions: denaturation at 94°C for 45 s, annealing at 60°C for 30 s and extension at 72°C for 45 s (28 cycles).
|
GENE |
PRIMER |
SEQUENCE |
|---|---|---|
|
CCL22/MDC |
Fw |
ACTGCACTCCTGGTTGTCCTC |
|
|
Rev |
CACGGTCATCAGTAGGCTC |
|
CCL17/TARC |
Fw |
ATGGCCCCACTGAAGATGCTGG |
|
|
Rev |
TCAAGACCTCTCAAGGCTTTGC |
|
CCL3/MIP-1α |
Fw |
CTGCTGCTTCAGCTACACCTCC |
|
|
Rev |
ACCCCTCAGGCACTCAGCTCC |
|
CCL5/RANTES |
Fw |
TATTCCTCGGACACCACAC |
|
|
Rev |
GCTCATCTCCAAAGAGTTGA |
|
CCL20/LARC |
Fw |
TTGCTCCTGGCTGCTTTGATG |
|
|
Rev |
TCTTTCTGTTCTTGGGCTATG |
|
CCL21/SLC |
Fw |
AGCCTCCTTATCCTGGTTCTG |
|
|
Rev |
TTACAAGGAAGAGGTGGGGTG |
|
CCL19/ELC |
Fw |
ATGGCCCTGCTACTGGCCCTC |
|
|
Rev |
GCTCCCTCTGCACGGTCATAG |
|
CCL1/I-309 |
Fw |
GTGGTGAGCTCTTAGCTTCACC |
|
|
Rev |
ACCAAGCAGATCCTCTGTGACC |
|
CCL25/TECK |
Fw |
AACCTGTGGCTCCTGGCCTGC |
|
|
Rev |
GGCTCACAGTCCTGAATTAGC |
|
CCL18/PARC |
Fw |
TGCCCTCCTTGTCCTCGTCTG |
|
|
Rev |
GAAGGGAAAGGGGAAAGGATG |
|
CXCL8/IL-8 |
Fw |
ATGACTTCCAAGCTGGCCGTGG |
|
|
Rev |
TCTGGCAACCCTACAACAGACC |
|
CXCL10/IP-10 |
Fw |
AACTGCGATTCTGATTTGCTG |
|
|
Rev |
TTGGAAGATGGGAAAGGTGAG |
|
CXCL9/MIG |
Fw |
TTGCTGGTTCTGATTGGAGTG |
|
|
Rev |
GTTGTGAGTGGGATGTGGTTG |
|
CXCL12/SDF-1 |
Fw |
ATGAACGCCAAGGTCGTGGTCG |
|
CXCL12/SDF-1α |
Rev |
AAGTCCTTTTTGGCTGTTGTGC |
|
CXCL12/SDF-1β |
Rev |
TGACCCTCTCACATCTTGAACC |
|
CXCL13/BCA-1 |
Fw |
GCCGCCACCATGAAGTTCATCTCGACATCTC |
|
|
Rev |
TAGTGGAAATATCAGCATCAGG |
|
CX3CL1/Fractalkine |
Fw |
CCACCTTCTGCCATCTGACTG |
|
|
Rev |
CCATCTCTCCTGCCATCTTTC |
|
Actin |
Fw |
GTGGGGCGCCCCAGGCACCA |
|
|
Rev |
CTCCTTAATGTCACGCACGATTC |
4.6 ELISA
The production of the chemokines CCL3, CCL5, CCL17, CCL22 and CXCL8 was quantified by ELISA using commercially available kits (R&D Systems, Minneapolis, MN for CCL22 and CCL17; BioSource International Inc., Camarillo, CA for CCL3 and CCL5 and Bender MedSystem, Vienna, AU for CXCL8). The lower limit of detection of the ELISA was 0.002 ng/ml for CCL3, 0.003 ng/ml for CCL5, 0.007 ng/ml for CCL17, 0.0625 ng/ml for CCL22, and 0.011 ng/ml for CXCL8, respectively.
4.7 Chemotaxis assay
Chemotaxis assays were performed using the Transwell system (5-μm pores, Costar, Cambridge, MA), as previously described 38, 39. Cell supernatants obtained from leukemia cells cultured with and without CD40L (500 μl) were thawed and used at 100% vol/vol. Recombinant CCL22 (250 and 500 ng/ml) (R&D Systems, Minneapolis, MN) was added to cRPMI anddistributed on cluster plates and warmed for 15 min at 37°C. Following incubation with culture medium, the Transwell inserts were transferred into the pre-warmed cluster plates. CCR4+ CD4+ recently activated T cells (5×105 cells for 6.5 mm inserts) were placed into the Transwell inserts. Plates were incubated for 1 h at 37°C, 5% CO2. In parallel experiments, neutralizing anti-CCL22 antibodies (10 μg/ml, R&D) were added to the supernatants, prior to the chemotaxis assay.
All migrated cells were collected from the lower compartment and counted by flow cytometry. The flow cytometer settings were determined prior to the acquisition of the migrated cells using samples of the input population. The migration percentage was calculated by dividing the number of migrated cells by the number of input cells.
4.8 Immunohistochemistry
Frozen sections were obtained from snap-frozen fragments of LN (four samples) and BM trephine biopsies (three samples) from patients with CLL. To evaluate the relationship between proliferation centers and T cells we investigated LN samples with double-marker immunostaining technique, 40. Ki67-reactive cells, clustering in pseudo-follicles, were revealed together with CD4-expressing lymphocytes. Cryostat serial sections of LN were transferred onto glass slides covered with polylysine adhesive, fixed in chloroform-acetone 1:1 mixture for 5 min and air dried. Ki67 antibody (Dako, Glostrup, Denmark) was incubated first at 1:100 dilution, and revealed using a sensitive avidin-streptavidin-peroxidase technique (Biogenex San Ramon, CA). After development of peroxidase with H2O2 and 3,3′-diaminobenzidine containing 1% nickel-chloride, the preparations were thoroughly washed in PBS, incubated with anti-CD4 mAb 1:20 dilution (Dako) and revealed using an alkaline-phosphatase anti-alkaline phosphatase immunostaining technique (APAAP, Dako) with new-fuchsin development providing red immunostaining of CD4+ cells. The same technique was used to demonstrate in the same section Ki67 and CD40L, but the signal of the latter marker was unreliable.
To reveal CD40L+ cells, sections were fixed in cold acetone/chloroform 1:1 mixture for 1 min, and washed with PBS before immunostaining. CD40L antibody (clone TRAP1, Pharmingen-BectonDickinson) was used at 1:50 dilution in PBS for 30 min at room temperature. An avidin-streptavidin peroxidase immunoperoxidase system (Stravigen multilink kit, Biogenex) was used as detection system, following manufacturer's instructions.
Acknowledgements
GS is a recipient of a fellowship from "Gruppo di Cooperazione in Cancerologia". We gratefully acknowledge Ms. Cristina Scielzo's technical experience.This work was supported by the "Associazione Italiana per la Ricerca sul Cancro" (AIRC), Milano, Italy, MURST 40%, 60% and by the MDACC Laboratory Study Agreement LS01–039.
- WILEY-VCH
- WILEY-VCH
- WILEY-VCH
- WILEY-VCH
- WILEY-VCH




