GL7 defines the cycling stage of pre-B cells in murine bone marrow
Abstract
We have identified a novel subset of early B lineage cells in the mouse bone marrow (BM) by GL7 expression on cell surface. GL7+B220low BM cells have a large cell size and are CD43–to low, CD95–, Sca-1–, I-Alow, IgM– and IgD–, suggesting that they are large pre-B cells. These BM cells express λ5 and VpreB but notterminal deoxytransferase (TdT) and Bcl-2, and approximately 50 % of them are in cell cycle. This fraction was not detected in BM cells of Rag-1-deficient and Scid mice, supporting that GL7+B220low BM cells belong to fraction C′ and D according to Hardy's criteria or to an early large pre-B-II fraction according to Melchers-Rolink's criteria. Furthermore, GL7+B220low BM cells can differentiate into IgM+ immature B cells in co-culture with stromal cells. These results suggest that B lymphocytes pass through the GL7+ pre-B cell stage during differentiation in the BM. Thus, GL7 is the critical marker to define the proliferation stage of large pre-B cells.
Abbreviations:
-
- BCR:
-
B cell receptor
-
- BM:
-
Bone marrow
-
- APC:
-
Allophycocyanin
-
- PI:
-
Propidium iodide
-
- TdT:
-
Terminal deoxytransferase
1 Introduction
When mice are immunized with T cell-dependent Ag, Ag-reactive mature B cells in the spleen are activated by interaction with CD4+ helper T cells in periarteriolar lymphoid sheaths, and migrate into follicles to form germinal centers 1, 2. Those germinal center B cells further differentiate into antibody-forming cells or memory B cells. Germinal center B cells are identified by their surface expression of a 35-kDa protein detected by mAb GL7 3. Although the number of GL7-positive B and T cells in the spleen of naive mice is very low (0 to 1 %), activation of B and T cells by various stimuli resulted in high levels of GL7 expression. GL7 thus appears to be a cell-surface molecule expressed selectively on subpopulations of activated B and T cells. In contrast, GL7 is also expressed on 1 – 2 % of B220+ cells in the bone marrow (BM) of naive adult mice and on a subpopulation comprising approximately 20 % of TCR-bright thymocytes, indicating expression of GL7 on intermediate stages of developing lymphocytes. However, GL7+B220+ BM cells have never been characterized on the basis of acareful delineation of early B lineage cells.
B lineage cells develop in the liver during embryonic life and in the BM after birth, and can be recognized by expression of the high molecular weight form of the CD45R (B220) 4, 5. Developmental stages of B cells are primarily defined by rearrangement of the Ig H and L chain genes and by the absence or presence of specific surface markers 6. Hardy and colleagues 7 fractionated intermediate stages of developing B cells in the BM into seven fractions, based on differential expression of several cell surface molecules. The first committed population corresponds to pro-B cells that rearrange their V(D)J gene segments 8. They are large proliferating cells that express low level of B220 and high level of CD43 (fraction A – C). Fraction C is further subdivided into fraction C and C′ by the amount of CD24 expression. Furthermore, the majority of B220low CD43+CD24high (fraction C′) cells are in cell cycle 9. These B220lowCD43+ BM cells in fraction C-C′ express the IgM H chain in the cytoplasm and on the cell surface in association with surrogate L chains 10, and differentiate into pre-B cells (fraction D) that lose expression of CD43 and surrogate L chains. Following productive VJ gene rearrangements of the L chain, the pre-B cells give rise to virgin B cells (fraction E) that express IgM on their surface. Virgin B cells become IgM+IgD+ mature B cells (fraction F) that migrate into peripheral lymphatic organs. Here we characterized GL7+B220low cells in the BM of naive mice, and provide evidence that GL7+B220low BM cells belong to a subset of large pre-B cells.
2 Results
2.1 GL7+ BM B cells are distinct from GL7+ germinal center B cells
GL7+B220+ cells were identified in the BM of naive adult C57BL / 6 mice and their percentage was 1 – 2 % of total BM cells 3, 11. We examined the ontogeny of GL7+B220+ cells in fetal liver, newborn liver, and BM cells of mice until 4 weeks of age. GL7+B220+ cells became detectable in newborn liver (Fig. 1). Their percentage increased in BM cells and reached a maximum (3.1 %) at 2 weeks of age and decreased to 1.5 % at 4 weeks. These results suggest that GL7+B220+ BM cells belong to intermediate stages of developing B cells.
GL7+B220+ cells were also detected in the spleen of adult mice on day 14 after immunization (Fig. 2 A). However, the level of B220 on GL7+ BM cells of naive mice was lower than that on GL7+ splenic B cells of immunized mice. We compared surface phenotypes of GL7+B220low BM cells of naive mice with those of GL7+ splenic B cells of immunized mice (Fig. 2 B). GL7+ splenic B cells were CD43+ / –, CD95+, Sca-1+, I-A+, IgM+ / – and IgD+/– that are typical surface markers of germinal center B cells 11. This surface phenotype was different from that of GL7+B220low BM cells. Those BM cells were CD43– to low, CD95–, Sca-1–, I-Alow, IgM–, IgD– (Fig. 2 B), c-kit– and CD25+ (data not shown), suggesting that this fraction belongs to early B cell lineage.
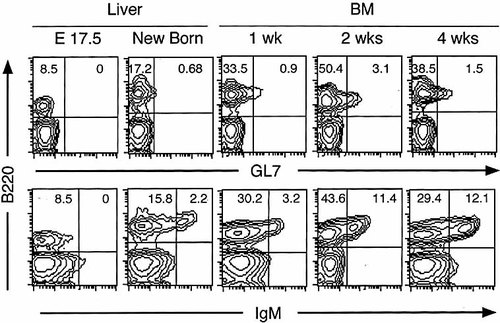
Ontogeny of GL7+B220+ BM cells. Fetal liver (E17.5), new born liver and BM cells of 1-, 2- and 4-week-old C57BL / 6J mice were stained with GL7-FITC, IgM-PE and B220-APC. The number in the corner indicates percentage of positively stained cells. A representative result from three to five mice.

Flow cytometric analysis of GL7+ B220+ cells in naive BM cells and immunized spleen cells. (A) BM cells of naive C57BL / 6J mice and spleen cells from DNP-OVA-immunized C57BL / 6J mice 12 days after immunization were stained with mAb B220-APC, GL7-FITC. (B) GL7+B220+ cells were further analyzed for expression of CD43, Fas, Sca-1, I-A, IgM and IgD. A representative result from three independent experiments.
2.2 GL7+B220low BM cells are early pre-B cells
The stages of B cell development in the BM can be identified by expression of stage-specific molecules 7, including TdT 12, λ5, and VpreB. To investigate the stages of GL7+B220low BM cells during B cell development, we sorted GL7+B220low cells, B220lowCD43+IgM– (fraction A – C′), B220+CD43–IgM– (fraction D) or B220+CD43–IgM+ (fraction E + F) fraction in Hardy's classification, splenic B (GL7–B220+) cells from naive mice, and germinal center B (GL7+B220+) cells from the spleen of immunized mice, and performed RT-PCR analysis (Fig. 3 A). Expression of TdT and the surrogate L chain genes is largely restricted to pro-B cells of fraction A – C′ 13, 14. Indeed, we detected high expression of TdT, λ5 and VpreB in BM cells of fraction A – C′), a lower (a 50-fold decrease) or undetectable expression of these markers in BM cells of fraction D and E + F. GL7+B220low BM cells expressed high levels of λ5, lower level of VpreB, and undetectable level of TdT. The VpreB protein was also weakly detected in the cytoplasm by immunohistochemistry but not on the cell surface of GL7+B220low BM cells by FACSCalibur (data not shown). Expression of Bcl-2 family is also distinct among those BM cells. Bcl-2 was detected in BM cells of fraction A – C′ and E + F but not in those of fraction D as previously reported 15. However, Bcl-2 was not detected in GL7+B220low BM cells.
Since pre-B (pre-B-II) cells are subdivided according to their cell size by Melchers-Rolink's fractionation 16 and L chain gene rearrangements are detected in small pre-B-II cells but not large pre-B-II cells 17, PCR amplifications of gene segments in two steps were used to assay the rearrangement status of the four κ L chain alleles and the λ L chain loci in GL7+B220low BM cells (Fig. 3 B). Single GL7+B220low BM cells were analyzed by PCR. Although about 95 % of all naive splenic B cells expressed κ L chain (data not shown), no κ and λ chain rearrangement was detected in GL7+B220low BM cells (20 single cells analyzed). These results suggest that GL7+B220low BM cells belong to late pro-B / early pre-B cells in fraction C-C′ 14 and large pre-B (pre-B-II) cells in fraction D.

RT-PCR analysis of gene expression and Ig L chain gene rearrangements in the B lineage cells. (A) B220lowCD43+IgM– (fraction A – C′), GL7+B220low, B220+CD43–IgM– (fraction D) and B220+CD43–IgM+ (fraction E + F) BM cells, splenic B cells from naive C57BL / 6J mice, and germinal center B cells (GL7+B220+ spleen cells from C57BL / 6J mice 12 days after DNP-OVA immunization) were sorted, and RT-PCR was performed. (B) Single GL7+B220low BM cells were isolated and single-cell PCR for κ and λ chain genes was performed. A representative result from 20 individual clones is shown.
2.3 GL7+B220low BM cells belong to a novel fraction of early B cell lineage
As Hardy's and Melchers-Rolink's fractionation of early B lineage cells were originally established in BALB / c mice, we analyzed expression of GL7 on BM cells of naive adult BALB / c mice (Fig. 4). Expression patterns of BALB / c mice were essentially identical to those of C57BL / 6 mice. Furthermore, the development arrest of pro-B cells to pre-B cells is demonstrated in Scid mice 18 and several genetically deficient mice lacking the H chain gene 19, the recombination activation (Rag-1 and Rag-2) genes 11, 20, the surrogate L chain gene 21. GL7+B220low cells were not detected in the BM of Rag-1-deficient (Fig. 4) and Scid (data not shown) mice. B cell development in the BM of those mice was arrested at fraction C (B220lowCD43+CD24low) 9, supporting the fact that GL7+B220low BM cells belong to more mature stages than late pro-B cells in fraction C.
The cell size of GL7+B220low BM cells was confirmed by flow cytometry. Fig. 5 shows that GL7+B220low BM cells were clearly larger than GL7–B220low and GL7–B220high BM cells. Furthermore, early pre-B cells in fraction C' 14 can be distinguished from late pro-B cells in fraction C by cell cycle analysis 9. We sorted GL7–B220high, GL7+B220low and GL7–B220low BM cells, and analyzed their cell cycle state. Approximately 45 % of GL7+B220low BM cells were in the S / G2M phase, whereas less than 5 % of GL7–B220high and GL7–B220low BM cells were in the S / G2M phase.
To confirm the presence of GL7+ cells in large pre-B cells, expression of GL7 was examined on BM cells in fraction A – C′ (B220lowCD43+IgM–) and fraction D (B220+CD43–IgM–) (Fig. 6 A). Approximately 20 % of large cells in fraction A – C′ were GL7+ (0.4 % of total BM cells), and small cells in fraction A – C′ did not express GL7 (Fig. 6 B). In addition, approximately 10 % of large cells in fraction D were GL7+ (1.4 % of total BM cells), and small cells in fraction D did not express GL7. These results indicate that GL7+B220low BM cells belong to a transitional stage from fraction C′ to fraction D by Hardy's criteria.
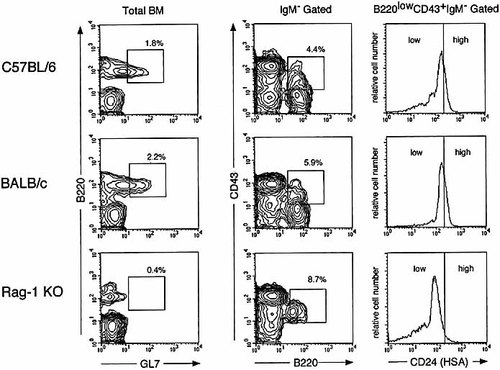
Expression of GL7 on BM cells from Rag-1-deficient mice. BM cells of C57BL / 6J, BALB / c and Rag-1-deficient (C57BL / 6J background) mice were stained with B220-APC and GL7-FITC, B220-APC, IgM-biotin / avidin-PerCP, CD43-PE and CD24(HSA)-FITC. A representative result from three independent experiments.
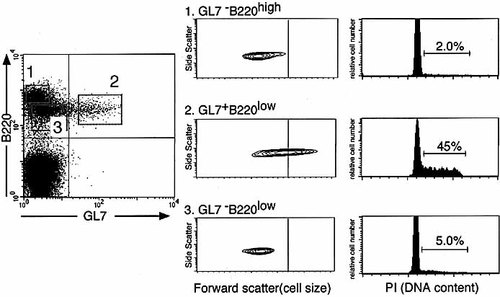
Cell size and cell cycle analysis of GL7+B220low BM cells. GL7–B220high (1), GL7+B220low (2) and GL7– B220low (3) BM cells of naive C57BL / 6J mice were sorted by FACS. Forward scatter (cell size) profiles of the electronically gated subsets are shown. The cells were lysed and the nuclei were stained with PI. The DNA content in the nuclei was determined by FACS. The percentage of PI-labeled nuclei in the S / G2 / M phase of the cell cycle is indicated. A representative result from three independent experiments.
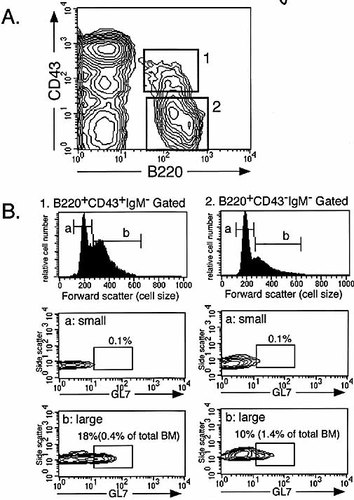
Expression of GL7 on large pre-B cells. (A) BM cells of naive C57BL / 6J mice were stained with B220-APC, IgM-biotin / avidin-PerCP, CD43-PE, and GL7-FITC. (B) Forward scatter of B220+CD43+IgM– BM cells (square 1 in Fig. 6 A) and B220+CD43–IgM– BM cells (square 2 in Fig. 6 A) were analyzed and divided by cell size. Expression of GL7 on those fractions (small cells, a; large cells, b in Fig. 6 B) was analyzed by flow cytometry. A representative result from three independent experiments.
2.4 GL7+B220low BM cells can differentiate into IgM+ immature B cells
In early B cell development, deletion by apoptosis occurs at the transition from pro-B to pre-B cell stage. As many as 75 % of developing B cells are lost at this stage of development 22. Since GL7+B220low BM cells belong to a transitional stage from fraction C′ to fraction D and did not express Bcl-2 (Fig. 3 A), these cells may be eliminated by apoptosis during maturation. In order to examine the possibility, we sorted GL7–B220lowCD43+IgM– BM cells and GL7+B220low BM cells, and further cultured the sorted cells on an OP-9 stromal cell layer with rIL-7 for 3 days (Fig. 7). When GL7–B220lowCD43+IgM– BM cells were cultured 33 % and 18 % became GL7+B220low and B220+IgM+ cells on day 3, respectively. In the culture of GL7+B220low BM cells, GL7+B220low cells proliferated and differentiated into GL7–B220+ cells and B220+IgM+ cells increased from 0 % to 59 % on day 3. These results indicate that GL7+B220low BM cells are able to differentiate into IgM+ B cells.
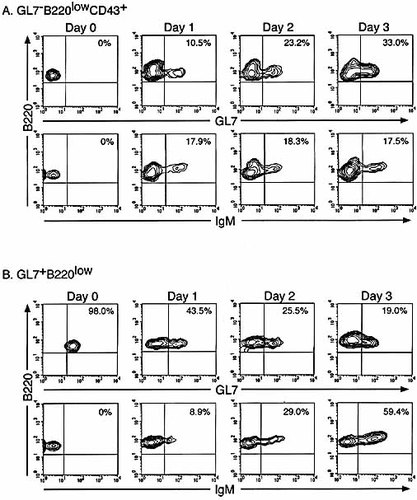
In vitro differentiation of GL7–B220lowCD43+ and GL7+B220low BM cells. Sorted GL7–B220lowCD43+ (A) and GL7+B220low (B) BM cells of naive C57BL / 6J mice were cultured over a confluent layer of OP-9 stromal cells supplemented with rIL-7 for 3 days. Expression of B220, IgM and GL7 on cultured BM cells was examined for 3 days. A representative result from three independent experiments.
3 Discussion
Developmental stages of B cells in the murine BM have been well defined by several groups. Osmond et al. have built up the simple schema of B cell development based on the TdT expression, cytoplasmic and surface μ expression, and cell size 23. Since GL7+B220low BM cells do not express TdT or surface IgM, and have a large cell size, they are large pre-B cells. Hardy and colleagues defined the developmental stages according to surface markers 7, with the advantage that each stage is defined only by flow cytometry and that living cells can be handled. According to Hardy's fractionation, GL7+B220low BM cells belong to early pre-B cells in fraction C′ and D. Although Hardy's and Osmond's classifications show some discrepancy especially in the borderline between pro-B cells and pre-B cells 24, GL7+B220low BM cells belong to the transitional stage of pro-B cells to pre-B cells by their criteria. On the other hand, Melchers et al. initially divided the stages of B cell development based on the changing status of Ig gene rearrangements, and a number of molecular phenotypic markers have added to their definition 25. GL7+B220low BM cells are defined as the early stage of large pre-B-II cells according to Melchers-Rolink's criteria. As μ expression and Ig rearrangement proceed in parallel with B cell maturation, these two classification are essentially identical 26. Thus, regardless of the classification of early B cell lineage, GL7+B220low BM cells belong to the fraction of early B cell lineage at the transitional stage of pre-B cells just after thepro-B cell stage.
GL7 was initially described as a mAb that reacts with a 35-kDa protein on subsets of activated T and B cells and germinal center B cells 3. Germinal center B cells undergo affinity maturation of their B cell receptors (BCR) and Ig isotype-switch, resulting in the formation of memory B cells 1, 2. Germinal center B cells are selected by their Ag specificity 27, and anti-apoptotic signals provided by follicular dendritic cells are crucial in this selection process. GL7 is also detected on 20 % of CD3high thymocytes 3. Those thymocytes are positively selected in the thymus 28. These results suggest that GL7 is an activation marker on developing B and T cellsthat successfully differentiate into further stages after the selection process by apoptosis. GL7+B220low BM cells are at the transitional stage of pre-B cells just after the pro-B cell stage. At this transitional stage, B cells progressively lose the contact to stromal cells and down-regulate the IL-7R 29, they exclusively depend on the pre-BCR for the last phases of development and enter a rapid proliferating stage. Since most of GL7+B220low BM cells were in cell cycle, GL7 may define the pre-BCR-dependent proliferation stage of large pre-B cells. Furthermore, GL7– pro-B cells express GL7 during differentiation to IgM+ immature B cells and GL7+B220low BM cells can differentiate into IgM+immature B cells in short-term stromal-dependent culture (Fig. 7), indicating that pre-B cells express GL7 during the normal differentiation pathway in the BM. Thus, GL7+B220low BM cells may be large pre-B cells that proliferate by the pre-BCR stimulation and successfully differentiate into small pre-B cells.
4 Materials and methods
4.1 Mice, Ag and immunization
C57BL / 6J, BALB / c and Scid mice were obtained from Clea (Tokyo). Rag-1-deficient mice were described previously 30. DNP-OVA was prepared by coupling of OVA (Sigma Chemical Co., St. Louis, MO) with 2,4-dinitrophenylbenzen-sulfonic acid under alkaline condition 31. Mice were immunized i. p. with 100 μg of alum-precipitated DNP-OVA, and their spleen cells were collected 14 days after immunization.
4.2 mAb and FACS analysis
Biotinylated, FITC-, PE- or allophycocyanin (APC)-conjugated mAb against B220 (RA3-6B2), CD43 (S7), CD24 (HSA; J11d), BP-1 (Ly-51; 6C3), IgM (R6-60.2), CD5 (Ly-1; 53-7.3), GL7 (Ly-77), FAS (Jo2), I-A (Aαb), Sca-1 (Ly 6A / E), or CD25 (IL-2Rα) were purchased from PharMingen (San Diego, CA). Biotinylated mAb were visualized using streptavidin-APC or streptavidin-PerCP (PharMingen). Cell suspensions were treated with lysing buffer (ACK; 0.155 M ammonium chloride, 0.1 M disodium EDTA, 0.01 M potassium bicarbonate) to lyse erythrocytes before staining. Single-cell suspensions were prepared in staining medium (PBS with 3 % FCS and 0.1 % sodium azide), and were stained with the mAb described above. After 20-min incubation on ice, cells were washed twice with staining medium and resuspended in staining medium supplemented with propidium iodide (PI, 1 μg / ml). Stained cells were analyzed on a FACSCalibur (Becton Dickinson, San Jose, CA). B lineage cells were resolved into various fractions (A – F) based on the technique initially reported by Hardy et al. 8, 9. Briefly, fraction A – C′ (B220+CD43+IgM– cells) were distinguished from fraction D (B220+CD43–IgM– cells) by staining to CD43, and fraction E + F cells were defined as B220+CD43–IgM+ cells.
4.3 Isolation of B220lowCD43+ or GL7+B220low BM cells and co-culture with stromal cells
Total BM cells were stained with GL7-FITC, CD43-PE, B220-APC for 20 min at 4 °C. After washing three times with staining medium, the cells were resuspended in staining medium supplemented with PI (1 μg / ml). Stained cells were analyzed by FACS Vantage (Becton Dickinson), and B220lowCD43+ or GL7+B220low BM cells were sorted. Murine stromal cell line, OP-9 32 was obtained from RIKEN cell bank (Tsukuba, Japan). OP-9 was seeed in 6-well plates (Becton Dickinson Labware, Lincoln Park, NJ) 1 day before co-culture. The sorted B220lowCD43+ or GL7+B220low BM cells were cultured on the OP-9 stromal layer with 3 ml of RPMI1640 containing murine rIL-7 (20 U / ml, kindly provided by Dr. TetuoSudo, Toray industries, Kamakura, Japan), 10 % heat-inactivated FCS, and 5 × 10– 5 M 2-ME 33.
4.4 Single-cell PCR analysis
Cells were sorted using FACS Vantage with Clon-cyte (Becton Dickinson). Single cells were directly sorted into 96-well plates (Falcon) containing 10 μl of 10 mM Tris-HCl (pH 8.3) per well.To prepare DNA, the samples were digested with 2 μl of proteinase K (5 mg / ml; Boehringer Mannheim, Mannheim, Germany) for 1 h at 55 °C followed by a 10-min incubation at 95 °C to inactive proteinase K. Plates were then stored at − 80 °C until used. DNA amplification of the rearranged L chain genes was carried out in two rounds of PCR using a TGradient Thermocycler (Whatman, Biometry, GB). The primer designs for the first- and second-round PCRs were previously described 17, 34. The following primers were used: universal Vκ, 5′-GGCTGCAGSTTCAGTGGCAGTGG(A / G)TC(A / T)GG(A / G)AC-3′; 5′ of Jκ1 (first round), 5′-GCTACCCACTGCTCTGTTCCTCTTCAGTG-3′; 5′ of Jκ1 (second round), 5′-TGTACAGCCAGACAGTGGAGTACTACCAC-3′; 3′ of Jκ1 (first round), 5′-CACTGGCATTCATTCTCCAGAGAACATGTC-3′; 3′ of Jκ1 (second round), 5′-TGCCTTGGAGAGTGCCAGAATCTGG-3′; 3′ of Jκ2 (first round), 5′-CAAAACCCTCCCTAGGTAGACAATTATCCCTC-3′; 3′ of Jκ2 (second round), 5′-GGACAGTTTTCCCTCCTTAACACCTGATCTG-3′;3′ of Jκ4 (first round), 5′-CTTCTACATTCCCCTTACAAAACTGGTCTC-3′; 3′ of Jκ4 (second round), 5′-TGATGCACAGGTTGCCAGGAATGG-3′; 3′ of Jκ5 (firstround), 5′-CCTCTCAACTAAAGCCTCTTTTTGCCCCTAATC-3′; 3′ of Jκ5 (second round), 5′-TGCCACGTCAACTGATAATGAGCCCTCTCC-3′; RS (first round) 5′-CATAACTGACTGTGCTGGCTGGGTTGG-3′; RS (second round) 5′-CTGCCCACACGACTCCTTCAGGCAGACG-3′; Vλ (first round) 5′-GCCATTTCCCAGGCTGTTGTGACTCAGG-3′; Vδ (second round) 5′-AATCTGCACTCACCACATCACCTGGTG-3′; and universal Lλ 5′-ACTCACCTAGGACAGTCAGCTTGGTTCC-3′. Briefly, the λL chain gene was assayed by a universal Vλ primer and a universal Jλ primer in the first step and a nested Vλ primer together with the universal Jλ primer in the second step. A Jλ primer used in these experiments binds equally well to Jλ1, Jλ2 and Jλ3. The κ chain gene rearrangements were assayed as described previously, and were extended to include Vκ rearrangements to the RS element. Efficiencies of all PCR reaction detecting single VκJκ1, VκJκ2, VκJκ4, VκJκ5 and VκRS rearrangements were near 90 %. The amplified DNA was analyzed by agarose gel electrophoresis and stained with ethidium bromide.
4.5 Cell cycle analysis
Cell cycle analysis was performed as described by Nicoletti et al. 35. Briefly, sorted cells were incubated in hypotonic lysing buffer (0.1 % sodium citrate, 0.01 % Triton X, 0.1 mg / ml RNase, and 0.1 mg / ml PI). DNA content in each nuclei was analyzed on FACSCalibur (Becton Dickinson, Mountain View, CA) using Cell Quest software for Macintosh.
4.6 RT-PCR analysis
Total RNA was extracted from BM cells using Trizol reagent (Life Technologies, Grand Island, NY). RNA were reverse-transcribed using Superscript II (Life Technologies) and oligo(dT) (Pharmacia, Piscataway, NJ), in a final volume of 20 μl, and 1 μl of cDNA was used for PCR. RT-PCR assays were carried out using the primer intially reported by Sigvardsson et al. 36. The primers used were as follows: β-actin 5′-GTTTGAGACCTTCAACACC-3′, 5′-GTGGCCATCTCCTGCTCGAAGTC-3′; Vpre B 5′-CGTCTGTCCTGCTCATGCT-3′, 5′-ACGGCACAGTAATACACAGCC-3′; λ5 5′-TGTGAAGTTCTCCTCCTGCTG-3′, 5′-ACCACCAAAGTACCTGGGTAG-3′; mb-1 5′-GCCAGGGGGTCTAGAAGC-3′, 5′-TCACTTGGCACCCAGTACAA-3′; TdT 5′-GAAGATGGGAACAACTCGAAGAG-3′, 5′-TGGCAGAGATTTCAGTACAGAGG-3′; bcl-xL 5′-TGGTCGACTTTCTCTCCTAC-3′ and 5′-AGAGATCCACAAAGATGTCC-3′ 37, bcl-2 5′-TCGCTACCGTCGTGACTTC-3′ and 5′-AAACAGAGGTCGCATGCTG-3′ 37, bax 5′-ACAGATCATGAAGACAGGGG-3′ and 5′-CAAAGTAGAAGAGGGCAACC-3′ 37. The PCR products were separated ona 1.5 % agarose gel and stained with ethidium bromide.
Acknowledgements
We would like to extend our thanks to Drs. J. Wang, M. Cooper and T. Sudo for their helpful discussion and collaboration, H. Satake for her technical assistance, and K. Ujiie for her secretarial assistance. This work was supported in part by the Grants-in-Aid from the Ministry of Education, Science, Technology, Sports and Culture of Japan.
- WILEY-VCH
- WILEY-VCH
- WILEY-VCH
- WILEY-VCH
- WILEY-VCH
- WILEY-VCH
- WILEY-VCH




